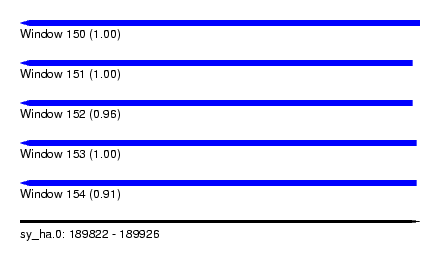
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 189,822 – 189,926 |
| Length | 104 |
| Max. P | 1.000000 |

| Location | 189,822 – 189,926 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.00 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 189822 104 - 2685015/80-200 CGAAGUAAAGUAAGCUCCUUAGCGCCGGUGGUAGCAGAUUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA-AUUAUU---------------GAGACCGAAAGGUCUCUUUUUUUAU ........((((((((....)))(((..(((((((.(.....)((((..(....)..))))))))))))))..-.)))))---------------((((((....))))))......... ( -32.90) >sy_au.0 534306 100 + 2809422/80-200 C-AAGUCAAGAAAGGUCUUUAGCGACGAUGGUAGCCAACUUACGUUCCGCUAGAGUAGAACGUUGCCAGGCAA-GUU------------------GAGACCGUGAGGUCUCUUUUUUUAU .-.....((((((((((......))).......(((.....((((((..(....)..)))))).....)))..-...------------------((((((....))))))))))))).. ( -30.80) >sy_sa.0 846800 120 + 2516575/80-200 CAUGAUAGGAAAAGCUCCUUAGCGCUGAUGGUAGCCGACUUACGUUCCGCAAGAGUAGGACGUUGCCAGGCAGCAUUAAUAGGAUGCGAUGAGCCGAGACCGAGAGGUCUCUUUUUUUAU ....((((((((((((((((((.((((.(((((((.(.(((((....(....).))))).))))))))..)))).)))..)))).))........((((((....))))))))))))))) ( -43.90) >consensus C_AAGUAAAGAAAGCUCCUUAGCGCCGAUGGUAGCCGACUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA_AUUA_U_______________GAGACCGAGAGGUCUCUUUUUUUAU ....(((((((((..........(((..(((((........((((((..(....)..))))))))))))))........................((((((....))))))))))))))) (-27.92 = -28.60 + 0.68)

| Location | 189,822 – 189,924 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.37 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -20.34 |
| Energy contribution | -21.23 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.65 |
| SVM decision value | 7.05 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 189822 102 - 2685015/120-240 UACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA-AUUAUU---------------GAGACCGAAAGGUCUCUUUUUUUAUGCCUAAAAUUAA--AACCGAAGGUUUAAUAAGAUAUCAAC .((((((..(....)..))))))....((((((-(..(..---------------((((((....))))))..)..)).))))).......(--((((...))))).............. ( -30.00) >sy_au.0 534306 89 + 2809422/120-240 UACGUUCCGCUAGAGUAGAACGUUGCCAGGCAA-GUU------------------GAGACCGUGAGGUCUCUUUUUUUAUGUCUAAAACGUC--AA----------AAUAAAAAGUAAAC (((((((..(....)..))))(((...((((((-(..------------------((((((....))))))..))....)))))..)))...--..----------........)))... ( -25.20) >sy_sa.0 846800 120 + 2516575/120-240 UACGUUCCGCAAGAGUAGGACGUUGCCAGGCAGCAUUAAUAGGAUGCGAUGAGCCGAGACCGAGAGGUCUCUUUUUUUAUGUCAAAAAAGGCUUAACUUCUCCAGUUCGAAGAACUGAAA .((((((..(....)..)))))).(((.(((.(((((.....))))).....)))((((((....))))))..................)))..........((((((...))))))... ( -38.90) >consensus UACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA_AUUA_U_______________GAGACCGAGAGGUCUCUUUUUUUAUGUCUAAAAAGAC__AAC________UAAUAAGAAAUAAAC .((((((..(....)..)))))).....((((.............((.....)).((((((....))))))........))))..................................... (-20.34 = -21.23 + 0.89)

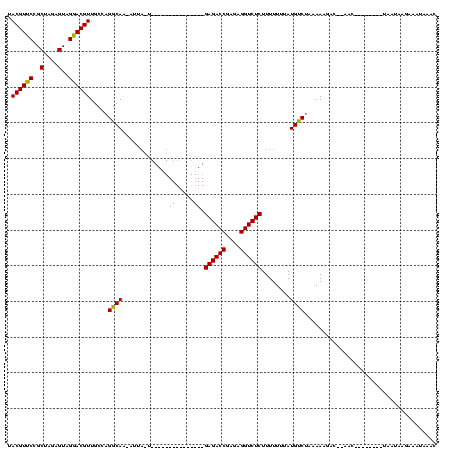
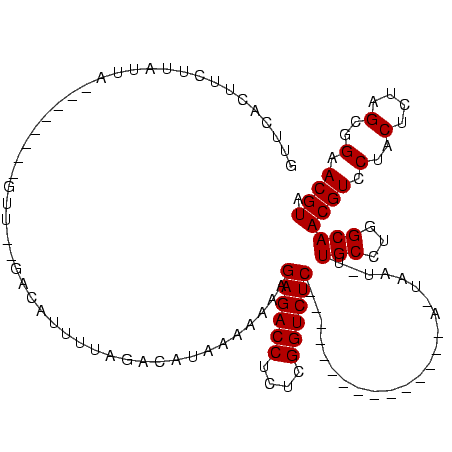
| Location | 189,822 – 189,924 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.37 |
| Mean single sequence MFE | -26.55 |
| Consensus MFE | -13.23 |
| Energy contribution | -13.23 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 189822 102 - 2685015/120-240 GUUGAUAUCUUAUUAAACCUUCGGUU--UUAAUUUUAGGCAUAAAAAAAGAGACCUUUCGGUCUC---------------AAUAAU-UUGCCUGGCAACGUCCUACUCUAGCGGAACGUA ((((.......((((((.(....).)--))))).(((((((........((((((....))))))---------------......-.))))))))))).(((.........)))..... ( -25.16) >sy_au.0 534306 89 + 2809422/120-240 GUUUACUUUUUAUU----------UU--GACGUUUUAGACAUAAAAAAAGAGACCUCACGGUCUC------------------AAC-UUGCCUGGCAACGUUCUACUCUAGCGGAACGUA ......((((((((----------((--((....))))).)))))))..((((((....))))))------------------...-.(((...)))((((((..(....)..)))))). ( -21.00) >sy_sa.0 846800 120 + 2516575/120-240 UUUCAGUUCUUCGAACUGGAGAAGUUAAGCCUUUUUUGACAUAAAAAAAGAGACCUCUCGGUCUCGGCUCAUCGCAUCCUAUUAAUGCUGCCUGGCAACGUCCUACUCUUGCGGAACGUA (((((((((...))))))))).......(((((((((......))))))((((((....))))))(((.....((((.......)))).))).))).((((((.........)).)))). ( -33.50) >consensus GUUCACUUCUUAUUA________GUU__GACAUUUUAGACAUAAAAAAAGAGACCUCUCGGUCUC_______________A_UAAU_UUGCCUGGCAACGUCCUACUCUAGCGGAACGUA .................................................((((((....)))))).......................(((...)))((((.(..(....)..).)))). (-13.23 = -13.23 + -0.00) # Strand winner: forward (1.00)

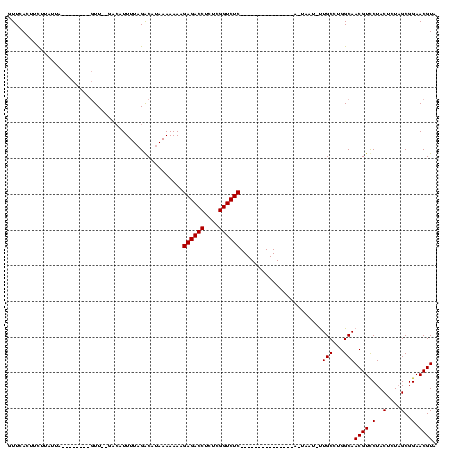
| Location | 189,822 – 189,925 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.14 |
| Mean single sequence MFE | -26.12 |
| Consensus MFE | -11.97 |
| Energy contribution | -12.97 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.46 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 189822 103 - 2685015/160-280 ---------------GAGACCGAAAGGUCUCUUUUUUUAUGCCUAAAAUUAA--AACCGAAGGUUUAAUAAGAUAUCAACAAAUCAAAGUGUAAUAGCUUUACGAGAAGAAAGCGUUUGA ---------------((((((....)))))).....((((((.........(--((((...))))).....(((........)))...))))))..(((((.(.....))))))...... ( -22.50) >sy_au.0 534306 90 + 2809422/160-280 ---------------GAGACCGUGAGGUCUCUUUUUUUAUGUCUAAAACGUC--AA----------AAUAAAAAGUAAACA---CAAAGAAAAAUGGCUUGGCGAAGUGAAAACGUUUGA ---------------((((((....)))))).................((((--((----------...............---..............))))))..((....))...... ( -18.45) >sy_sa.0 846800 114 + 2516575/160-280 AGGAUGCGAUGAGCCGAGACCGAGAGGUCUCUUUUUUUAUGUCAAAAAAGGCUUAACUUCUCCAGUUCGAAGAACUGAAAAGCCUAAAACAAGCUA---UUAUGA---GAAAGCGUUUGA .((((((..((((((((((((....)))))).((((((.....))))))))))))..(((((((((((...))))))...(((.........))).---....))---))).)))))).. ( -37.40) >consensus _______________GAGACCGAGAGGUCUCUUUUUUUAUGUCUAAAAAGAC__AAC________UAAUAAGAAAUAAACA___CAAAGAAAAAUAGCUUUACGA___GAAAGCGUUUGA ...............((((((....))))))......................................................................................... (-11.97 = -12.97 + 1.00)

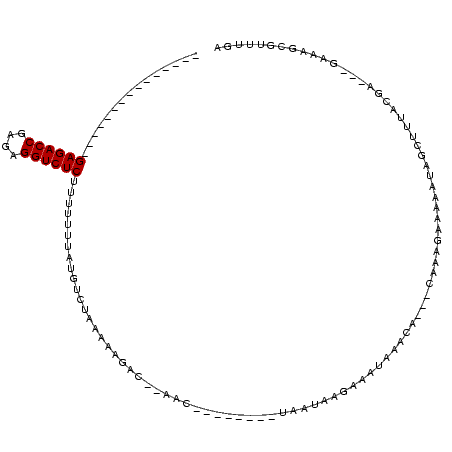
| Location | 189,822 – 189,925 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.14 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -8.97 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.41 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.912763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 189822 103 - 2685015/160-280 UCAAACGCUUUCUUCUCGUAAAGCUAUUACACUUUGAUUUGUUGAUAUCUUAUUAAACCUUCGGUU--UUAAUUUUAGGCAUAAAAAAAGAGACCUUUCGGUCUC--------------- ......((((..........(((.(((((.((........))))))).)))((((((.(....).)--)))))...)))).........((((((....))))))--------------- ( -17.50) >sy_au.0 534306 90 + 2809422/160-280 UCAAACGUUUUCACUUCGCCAAGCCAUUUUUCUUUG---UGUUUACUUUUUAUU----------UU--GACGUUUUAGACAUAAAAAAAGAGACCUCACGGUCUC--------------- ..(((((((...........((((((........))---.))))..........----------..--)))))))..............((((((....))))))--------------- ( -14.70) >sy_sa.0 846800 114 + 2516575/160-280 UCAAACGCUUUC---UCAUAA---UAGCUUGUUUUAGGCUUUUCAGUUCUUCGAACUGGAGAAGUUAAGCCUUUUUUGACAUAAAAAAAGAGACCUCUCGGUCUCGGCUCAUCGCAUCCU ......((((((---((....---.((((((...))))))...((((((...))))))))))))...((((((((((......))))))((((((....))))))))))....))..... ( -32.80) >consensus UCAAACGCUUUC___UCGUAAAGCUAUUUUACUUUG___UGUUCACUUCUUAUUA________GUU__GACAUUUUAGACAUAAAAAAAGAGACCUCUCGGUCUC_______________ .........................................................................................((((((....))))))............... ( -8.97 = -8.97 + 0.00) # Strand winner: forward (0.94)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:43 2006