| Sequence ID | sy_sa.0 |
|---|---|
| Location | 846,508 – 846,623 |
| Length | 115 |
| Max. P | 0.999994 |

| Location | 846,508 – 846,623 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -32.41 |
| Energy contribution | -31.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 846508 115 + 2516575/80-200 GAACACAGAAGUUAAGCUCCUUAGCGCCGAUGGUAGUCGGACUUACGUUCCGCAAGAGUAGGACGUUGCCAGGCAG--CA--UUAAUAGGAUGCGAUGA-GCCGAGACCGAGAGGUCUCU ...............((((......(((..((((((.((..(((((....(....).))))).))))))))))).(--((--((.....)))))...))-)).((((((....)))))). ( -41.00) >sy_au.0 534023 117 + 2809422/80-200 GAACACAGAAGUUAAGCUCCUUAGCGUCGAUGGUAGUCGAACUUACGUUCCGCUAGAGUAGAACGUUGCCAGGCAA--AAAAUGGAUGCGAUGAGCCGC-AUUGAGACCGCAAGGUCUCU ...............((((....(((((.((....(((......((((((..(....)..)))))).....)))..--...)).)))))...))))...-...((((((....)))))). ( -38.80) >sy_ha.0 145838 120 - 2685015/80-200 GAACACAGAAGUUAAGCUCCUUAGCGCCGAUGGUAGUUGGAUUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAAAUCGUAUGAAUGCGAUGAGCCGAUAUCGAGACCGAAAGGUCUCU ..........((((((...))))))...((((.....(((....((((((..(....)..))))))..)))(((..((((((....))))))..)))..))))((((((....)))))). ( -42.90) >consensus GAACACAGAAGUUAAGCUCCUUAGCGCCGAUGGUAGUCGGACUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA__CA_AUGAAUGCGAUGAGCCGA_AUCGAGACCGAAAGGUCUCU ..........((((((...))))))(((..(((((.........((((((..(....)..)))))))))))))).............................((((((....)))))). (-32.41 = -31.97 + -0.44)

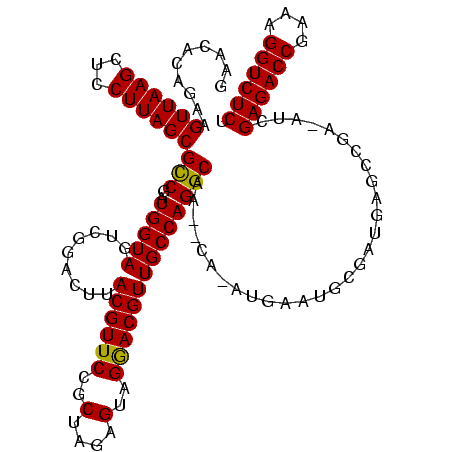
| Location | 846,508 – 846,618 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.60 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.62 |
| SVM decision value | 5.83 |
| SVM RNA-class probability | 0.999994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 846508 110 + 2516575/120-240 ACUUACGUUCCGCAAGAGUAGGACGUUGCCAGGCAG--CA--UUAAUAGGAUGCGAUGA-GCCGAGACCGAGAGGUCUCUUUUUUU-AUGUCAAAAAA----GGCUUAACUUCUCCAGUU .(((((....(....).)))))..(((((...))))--).--......(((...(.(((-(((((((((....)))))).((((((-.....))))))----)))))).)...))).... ( -34.50) >sy_au.0 534023 107 + 2809422/120-240 ACUUACGUUCCGCUAGAGUAGAACGUUGCCAGGCAA--AAAAUGGAUGCGAUGAGCCGC-AUUGAGACCGCAAGGUCUCUUUUUUUUAUGUCUAAAAC----GUCAAAAU------AAAA ....((((((..(....)..))))))....(((((.--((((..((((((......)))-)))((((((....))))))....)))).))))).....----........------.... ( -33.50) >sy_ha.0 145838 119 - 2685015/120-240 AUUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAAAUCGUAUGAAUGCGAUGAGCCGAUAUCGAGACCGAAAGGUCUCUUUUUUU-AUGCACAAAAACAAAACCGAAGGUUUAAUGAAA ....((((((..(....)..))))))(((..(((..((((((....))))))..)))......((((((....)))))).......-..)))........(((((...)))))....... ( -36.60) >consensus ACUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA__CA_AUGAAUGCGAUGAGCCGA_AUCGAGACCGAAAGGUCUCUUUUUUU_AUGUCAAAAAA____GCCAAAACUU____AAAA ....((((((..(....)..)))))).....((((..............(((........)))((((((....)))))).........))))............................ (-21.45 = -21.23 + -0.21)

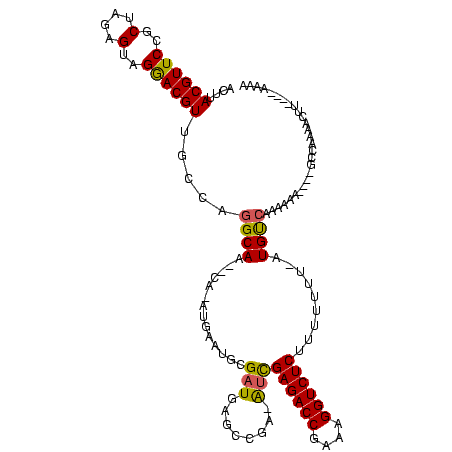
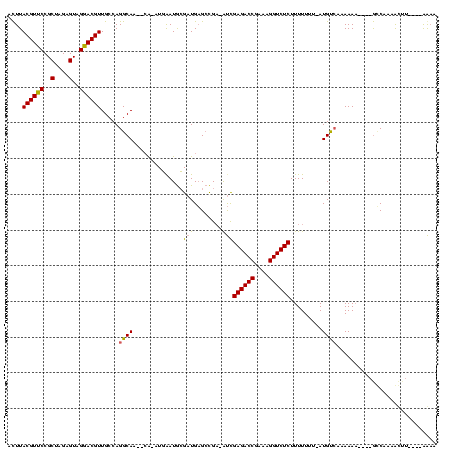
| Location | 846,508 – 846,619 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.26 |
| Mean single sequence MFE | -26.95 |
| Consensus MFE | -13.07 |
| Energy contribution | -13.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.48 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 846508 111 + 2516575/160-280 --UUAAUAGGAUGCGAUGA-GCCGAGACCGAGAGGUCUCUUUUUUU-AUGUCAAAAAA----GGCUUAACUUCUCCAGUUCGAAGAACUGAAAAGCCUAAAAUAAGCUAUU-AUGAGAAA --..............(((-(((((((((....)))))).((((((-.....))))))----))))))..(((((((((((...))))))...(((.........)))...-..))))). ( -31.60) >sy_au.0 534023 105 + 2809422/160-280 AAUGGAUGCGAUGAGCCGC-AUUGAGACCGCAAGGUCUCUUUUUUUUAUGUCUAAAAC----GUCAAAAU------AAAAAGCAAACACAAAGA----AAAAUGGCUUGGCGAAGUGAAA ........((..((((((.-...((((((....))))))((((((((.(((.......----........------.........))).)))))----))).))))))..))........ ( -25.26) >sy_ha.0 145838 116 - 2685015/160-280 UAUGAAUGCGAUGAGCCGAUAUCGAGACCGAAAGGUCUCUUUUUUU-AUGCACAAAAACAAAACCGAAGGUUUAAUGAAAUCAAUAAAUCAAAA---UGUAGUAGCUUUACGAAGAGUAA ......(((((((......))))((((((....)))))).......-..)))................((((((.((....)).))))))....---.......(((((....))))).. ( -24.00) >consensus _AUGAAUGCGAUGAGCCGA_AUCGAGACCGAAAGGUCUCUUUUUUU_AUGUCAAAAAA____GCCAAAACUU____AAAAAGAAAAAAUAAAAA___UAAAAUAGCUUAACGAAGAGAAA .......................((((((....))))))................................................................................. (-13.07 = -13.07 + 0.00)

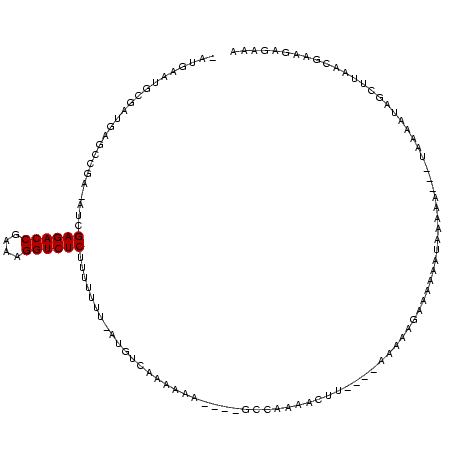
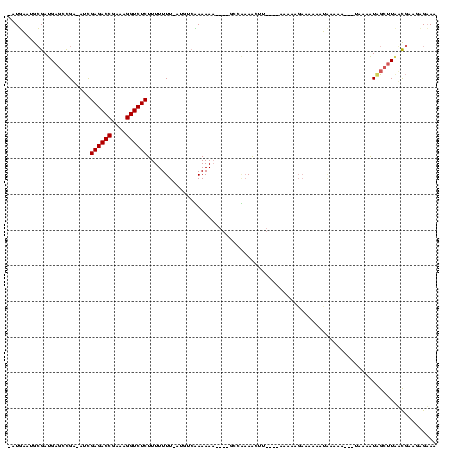
| Location | 846,508 – 846,619 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.26 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -10.10 |
| Energy contribution | -10.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 846508 111 + 2516575/160-280 UUUCUCAU-AAUAGCUUAUUUUAGGCUUUUCAGUUCUUCGAACUGGAGAAGUUAAGCC----UUUUUUGACAU-AAAAAAAGAGACCUCUCGGUCUCGGC-UCAUCGCAUCCUAUUAA-- .......(-(((((.........(((((((((((((...))))).)))))))).((((----((((((.....-.))))))((((((....)))))))))-).........)))))).-- ( -31.00) >sy_au.0 534023 105 + 2809422/160-280 UUUCACUUCGCCAAGCCAUUUU----UCUUUGUGUUUGCUUUUU------AUUUUGAC----GUUUUAGACAUAAAAAAAAGAGACCUUGCGGUCUCAAU-GCGGCUCAUCGCAUCCAUU .........((..((((..(((----(.((((((((((......------........----....)))))))))).))))((((((....))))))...-..))))....))....... ( -23.97) >sy_ha.0 145838 116 - 2685015/160-280 UUACUCUUCGUAAAGCUACUACA---UUUUGAUUUAUUGAUUUCAUUAAACCUUCGGUUUUGUUUUUGUGCAU-AAAAAAAGAGACCUUUCGGUCUCGAUAUCGGCUCAUCGCAUUCAUA ...........(((((....((.---....(.((((.((....)).)))).)....))...))))).((((..-.......((((((....))))))(((........)))))))..... ( -17.60) >consensus UUUCUCUUCGAAAAGCUAUUUUA___UUUUCAUUUAUUCAUUUU____AAAUUUAGAC____UUUUUAGACAU_AAAAAAAGAGACCUUUCGGUCUCGAU_UCGGCUCAUCGCAUUCAU_ .................................................................................((((((....))))))....................... (-10.10 = -10.10 + -0.00) # Strand winner: forward (0.90)

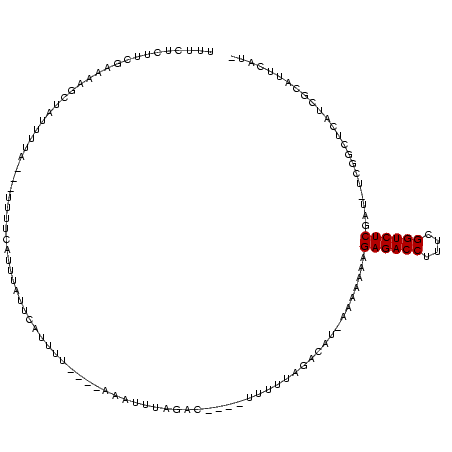
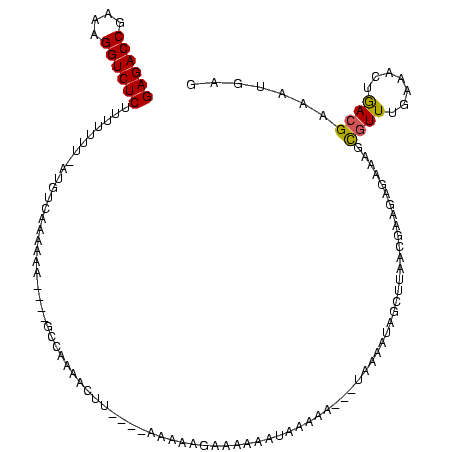
| Location | 846,508 – 846,622 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.94 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -13.17 |
| Energy contribution | -13.07 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.95 |
| SVM RNA-class probability | 0.999724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 846508 114 + 2516575/183-303 GAGACCGAGAGGUCUCUUUUUUU-AUGUCAAAAAA----GGCUUAACUUCUCCAGUUCGAAGAACUGAAAAGCCUAAAAUAAGCUAUU-AUGAGAAAGCGUUUGAAGCUGACGAACUGAG ((((((....)))))).......-.(((((..(((----.((((....((((((((((...))))))...(((.........)))...-..)))))))).))).....)))))....... ( -30.60) >sy_au.0 534023 106 + 2809422/183-303 GAGACCGCAAGGUCUCUUUUUUUUAUGUCUAAAAC----GUCAAAAU------AAAAAGCAAACACAAAGA----AAAAUGGCUUGGCGAAGUGAAAACGUUUGAAUCUGACGAAACGAG ((((((....))))))..................(----((((....------...((((...........----......))))..((((((....)).))))....)))))....... ( -23.63) >sy_ha.0 145838 116 - 2685015/183-303 GAGACCGAAAGGUCUCUUUUUUU-AUGCACAAAAACAAAACCGAAGGUUUAAUGAAAUCAAUAAAUCAAAA---UGUAGUAGCUUUACGAAGAGUAAGUGUUUAAAACUAAAGUAUUGAG ((((((....))))))...((((-(.((((...............((((((.((....)).))))))....---.......(((((....)))))..)))).)))))............. ( -26.50) >consensus GAGACCGAAAGGUCUCUUUUUUU_AUGUCAAAAAA____GCCAAAACUU____AAAAAGAAAAAAUAAAAA___UAAAAUAGCUUAACGAAGAGAAAGCGUUUGAAACUGACGAAAUGAG ((((((....))))))..................................................................................((((.......))))....... (-13.17 = -13.07 + -0.11)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:38 2006