
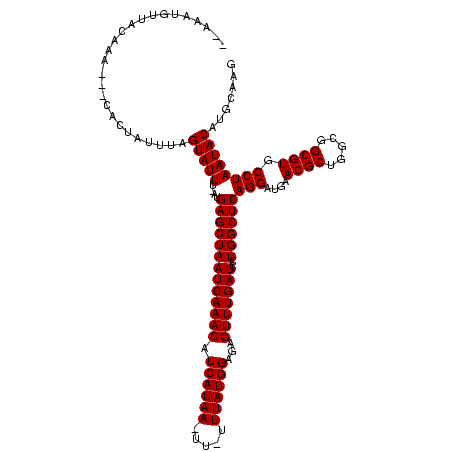
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 971,068 – 971,188 |
| Length | 120 |
| Max. P | 0.990710 |

| Location | 971,068 – 971,180 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -30.93 |
| Consensus MFE | -27.73 |
| Energy contribution | -27.73 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 112 + 2685015/80-200 --AAUUGUUACAAA---CACUAUUUAGUAUU--AUGAGCUAAUCAAACAUCAUAAAUU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAG --............---.........(((((--(((((((((((((((.((((((...-.))))))...)))))))..)))))))((....((((.....)))).))))))))....... ( -29.10) >sy_au.0 692085 111 - 2809422/80-200 --UAA-GUUACAAA---CAUUAUUUAGUAUUU-AUGAGCUAAUCAAACAUCAUAA-UU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAG --...-........---.........((((.(-(((((((((((((((.(((((.-..-..)))))...)))))))..))))))(((....((((.....)))).))).))).))))... ( -27.20) >sy_sa.0 841564 120 + 2516575/80-200 ACAAGUGUUAGAAAUAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAG ....((((((....))))))......(((((....(((((((((((((.((((((.....))))))...)))))))..))))))(((....((((.....)))).))))))))....... ( -36.50) >consensus __AAAUGUUACAAA___CACUAUUUAGUAUUU_AUGAGCUAAUCAAACAUCAUAA_UU_UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAG ..........................(((((....(((((((((((((.((((((.....))))))...)))))))..))))))(((....((((.....)))).))))))))....... (-27.73 = -27.73 + -0.00)

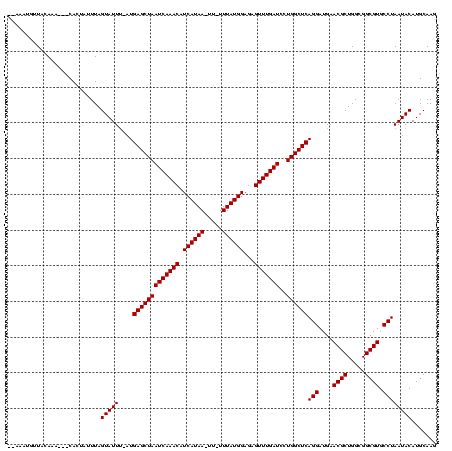
| Location | 971,068 – 971,180 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.80 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -23.60 |
| Energy contribution | -23.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 112 + 2685015/80-200 CUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA-AAUUUAUGAUGUUUGAUUAGCUCAU--AAUACUAAAUAGUG---UUUGUAACAAUU-- .(((((..((((((((.....)))...........(((((...((((((((....(((((.-...)))))..)))))))).))))).--.........)))))---..))))).....-- ( -26.90) >sy_au.0 692085 111 - 2809422/80-200 CUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA-AA-UUAUGAUGUUUGAUUAGCUCAU-AAAUACUAAAUAAUG---UUUGUAAC-UUA-- .(((((..((((((((.....)))...........(((((...((((((((....((((..-..-.))))..)))))))).))))).-..........)))))---..))))).-...-- ( -26.00) >sy_sa.0 841564 120 + 2516575/80-200 CUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAAUGAAUUAUGAUGUUUGAUUAGCUCAUAAAAUACUAAUUUGUGUUAUUUCUAACACUUGU .......((((((((.((((.....))))....)))((((...((((((((....(((((.....)))))..)))))))).))))....)))))......((((((....)))))).... ( -28.40) >consensus CUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA_AA_UUAUGAUGUUUGAUUAGCUCAU_AAAUACUAAAUAGUG___UUUGUAACAAUU__ .......((((((((.((((.....))))....)))((((...((((((((....(((((.....)))))..)))))))).))))....))))).......................... (-23.60 = -23.60 + -0.00) # Strand winner: forward (0.99)


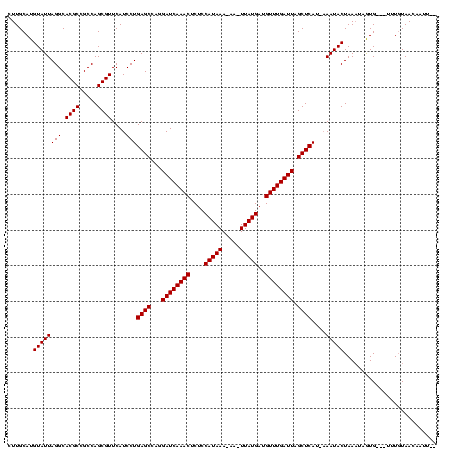
| Location | 971,068 – 971,187 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -36.79 |
| Energy contribution | -35.47 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 119 + 2685015/120-240 AAUCAAACAUCAUAAAUU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGACAAGGAGCUUGCUCCUUUGACGUUAGC .(((((((.((((((...-.))))))...))))))).(((((((((..((..(((((((.(((((.......)))))..))).))))..))..)((((((....))))))))).))))). ( -36.30) >sy_au.0 692085 118 - 2809422/120-240 AAUCAAACAUCAUAA-UU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACGGACGAGAAGCUUGCUUCUCUGAUGUUAGC .....(((((((...-..-..........(((((((((((...))))))...(((((((.(((((.......)))))..))).))))..)))))((((((....)))))))))))))... ( -38.90) >sy_sa.0 841564 120 + 2516575/120-240 AAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGAUAAGGAGCUUGCUCCUUUGACGUUAGC ..((((((.((((((.....))))))...))))))(((((...)))))....(((((((.(((((.......)))))..))).))))(((....((((((....))))))....)))... ( -35.10) >consensus AAUCAAACAUCAUAA_UU_UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGACAAGGAGCUUGCUCCUUUGACGUUAGC .(((((((.((((((.....))))))...))))))).(((((((((..((..(((((((.(((((.......)))))..))).))))..))..)((((((....))))))))).))))). (-36.79 = -35.47 + -1.33)

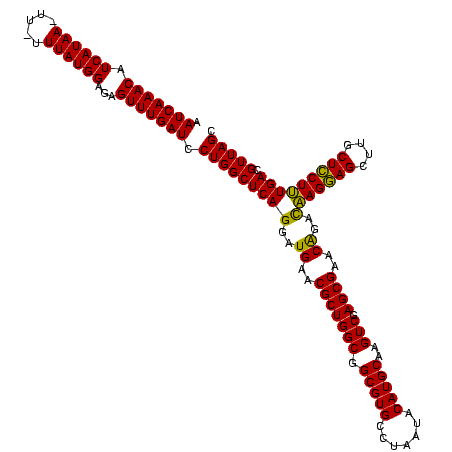

| Location | 971,068 – 971,187 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.15 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -31.71 |
| Energy contribution | -30.17 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 119 + 2685015/120-240 GCUAACGUCAAAGGAGCAAGCUCCUUGUCUGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA-AAUUUAUGAUGUUUGAUU ((.((((...((((((....))))))...)))).))..........((..(((((.((((.....))))....)))))..)).((((((((....(((((.-...)))))..)))))))) ( -31.00) >sy_au.0 692085 118 - 2809422/120-240 GCUAACAUCAGAGAAGCAAGCUUCUCGUCCGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA-AA-UUAUGAUGUUUGAUU .(.(((((((((((((....))))))((((....(((((...((.((((....(((.....))).)))).))...)))))..))))...............-..-...))))))).)... ( -32.20) >sy_sa.0 841564 120 + 2516575/120-240 GCUAACGUCAAAGGAGCAAGCUCCUUAUCUGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAAUGAAUUAUGAUGUUUGAUU .(.(((((((((((((....))))))....(((((((((...((.((((....(((.....))).)))).))...)))))..(((........)))......))))..))))))).)... ( -31.20) >consensus GCUAACGUCAAAGGAGCAAGCUCCUUGUCUGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA_AA_UUAUGAUGUUUGAUU .(.(((((((((((((....))))))((((....(((((...((.((((....(((.....))).)))).))...)))))..))))......................))))))).)... (-31.71 = -30.17 + -1.55) # Strand winner: forward (0.98)

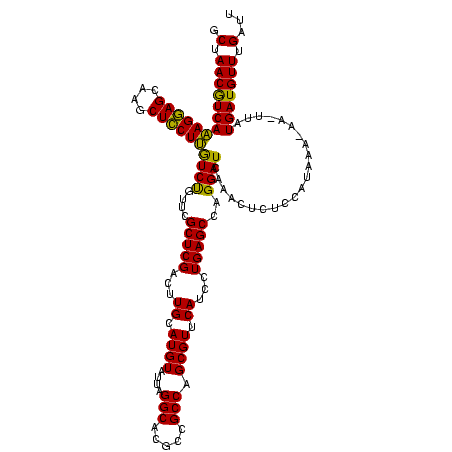
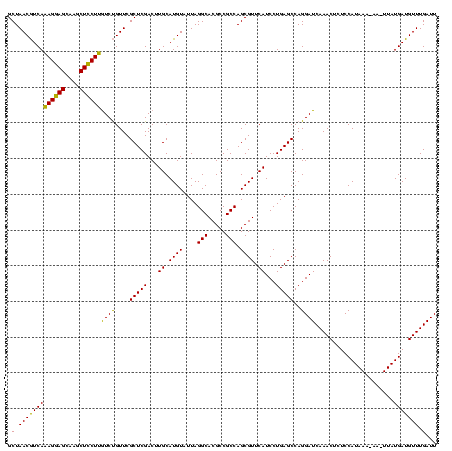
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -36.80 |
| Energy contribution | -36.47 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/280-400 ACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUG (((((.....((((((....))))))(((((((((((......))))...(((..((((((((((..(((.....)))..))))).(.....)..))))).)))..)))))))))))).. ( -40.10) >sy_au.0 692085 120 - 2809422/280-400 ACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUGAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUG (((((.....((((((....))))))(((((.((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))).)....)))))))))).. ( -38.60) >sy_sa.0 841564 120 + 2516575/280-400 ACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUAGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUG (((((.....((((((....))))))(((((((.((......((((((((......))))))))(((...(((((.....))))).((.......))...))).)))))))))))))).. ( -36.20) >consensus ACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCAAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUG (((((.....((((((....))))))(((((((.((......(((((((((....)))))))))(((...(((((.....))))).((.......))...))).)))))))))))))).. (-36.80 = -36.47 + -0.33)

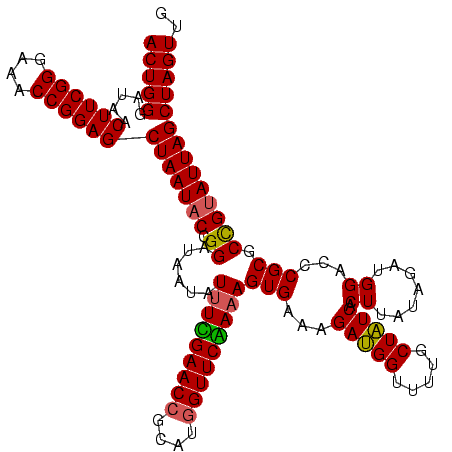
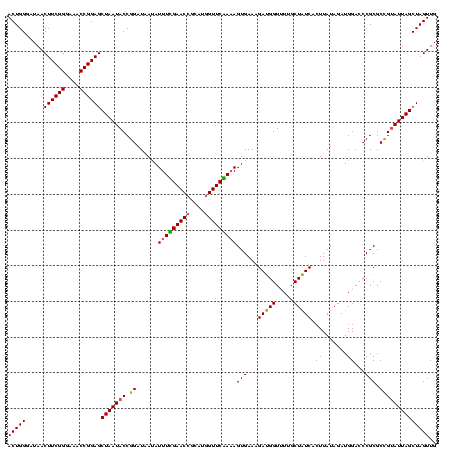
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -29.85 |
| Energy contribution | -29.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/280-400 CAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGU .(((((((((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......)))))))))))))).(((....))).))))........ ( -32.60) >sy_au.0 692085 120 - 2809422/280-400 CAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUCAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGU .(((((((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....)))))))))))))).(((....))).))))........ ( -33.10) >sy_sa.0 841564 120 + 2516575/280-400 CAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACUAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGU .(((((((((((((.(.((((((...((((.((((((.((.........)).))))))))))))).)))(((..(....)..)))).))))))))).(((....))).))))........ ( -26.90) >consensus CAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGU .(((((((((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......)))))))))))))).(((....))).))))........ (-29.85 = -29.30 + -0.55) # Strand winner: forward (1.00)

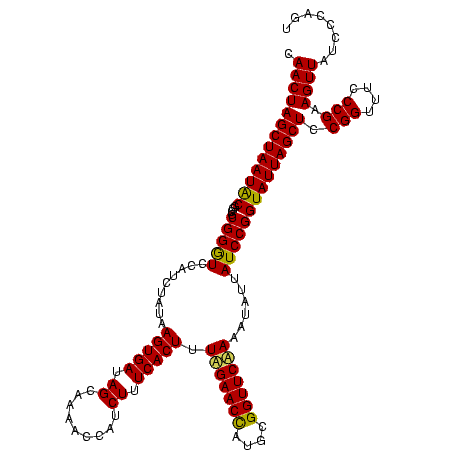
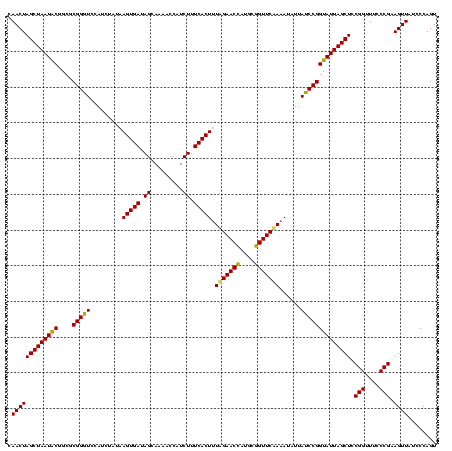
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -35.60 |
| Energy contribution | -34.27 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.06 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/320-440 UAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGAC ....(((((((....)))))))((((.....(((((....(((((.......)))))......)))))....)))).((((((((.......))))))))(((.(((...))).)))... ( -36.70) >sy_au.0 692085 120 - 2809422/320-440 UAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCAUAGCCGAC ..(((((((((....)))))))))((((..(((((.....)))))..)))).......((.((..((((((.(((..((((((((.......)))))))))))...))))))..)).)). ( -37.30) >sy_sa.0 841564 120 + 2516575/320-440 CAUUUGGAACCGCAUAGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGAC ..((((((((......))))))))((((..(((((.....)))))..))))..........((..((((((.(((..((((((((.......)))))))))))...))))))..)).... ( -32.40) >consensus UAUUUCGAACCGCAUGGUUCAAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGAC ..(((((((((....)))))))))((((..(((((.....)))))..))))..........((..((((((.(((..((((((((.......)))))))))))...))))))..)).... (-35.60 = -34.27 + -1.33)

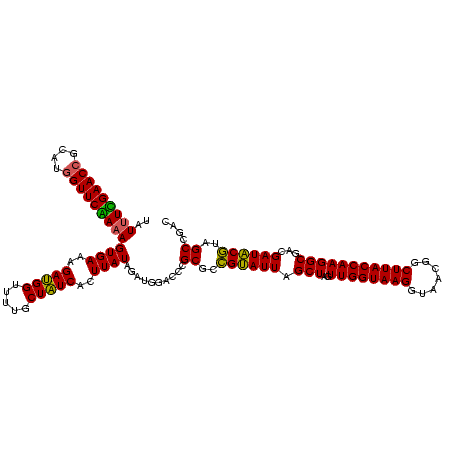
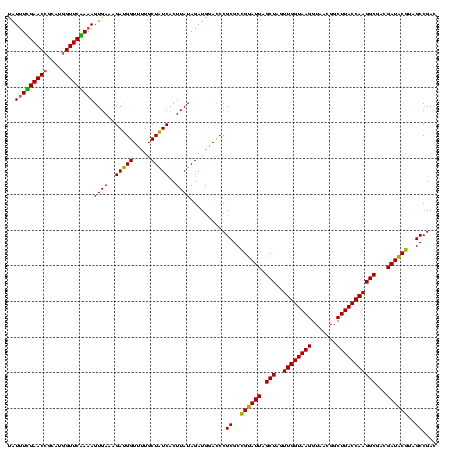
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -38.53 |
| Energy contribution | -42.87 |
| Covariance contribution | 4.34 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/480-600 CAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACA ....((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))... ( -45.90) >sy_au.0 692085 120 - 2809422/480-600 CAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACA ....((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))... ( -45.90) >sy_sa.0 841564 120 + 2516575/480-600 CAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGGUUUCGGCUCGUAAAACUCUGUUAUUAGGGAAGAACA ....((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))... ( -42.50) >consensus CAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACA .....((((((...........))))))..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))... (-38.53 = -42.87 + 4.34)

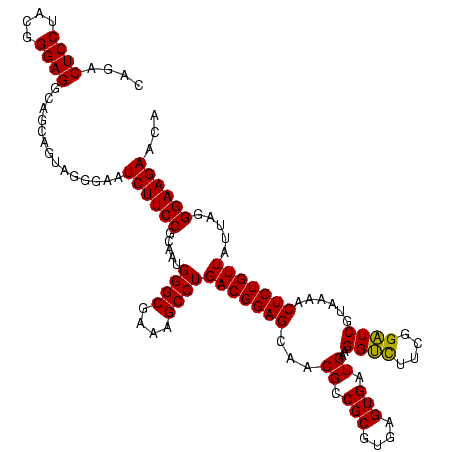
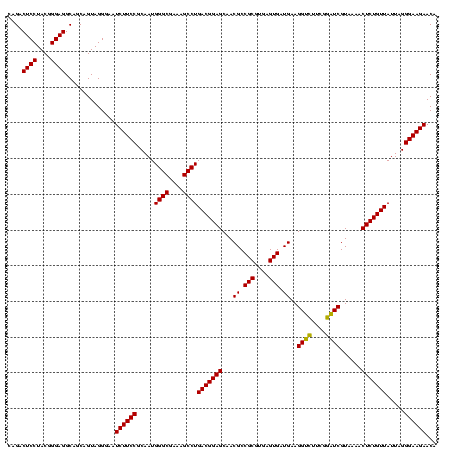
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -35.71 |
| Energy contribution | -39.60 |
| Covariance contribution | 3.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/520-640 UGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGUAACUAUGCACGUCUUGACGGUACCUAAUC .((((....))))(((((((...((.(((....))).))..((((....))))......)))))))((((((......((.(((((((........)))))))..))......)))))). ( -42.10) >sy_au.0 692085 120 - 2809422/520-640 UGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUAUGUGUAAGUAACUGUGCACAUCUUGACGGUACCUAAUC .((((....))))(((((((...((.(((....))).))..((((....))))......)))))))((((((......((.(((((((........)))))))..))......)))))). ( -40.70) >sy_sa.0 841564 120 + 2516575/520-640 UGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGGUUUCGGCUCGUAAAACUCUGUUAUUAGGGAAGAACAAACGUGUAAGUAACUGUGCACGUCUUGACGGUACCUAAUC .((((....))))(((((((...((.(((....))).))..((((....))))......)))))))((((((......((((((((((........))))))).)))......)))))). ( -39.50) >consensus UGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGUAACUGUGCACGUCUUGACGGUACCUAAUC .((((....))))(((((((...((.(((....))).))..((((....))))......)))))))((((((......((.(((((((........)))))))..))......)))))). (-35.71 = -39.60 + 3.89)

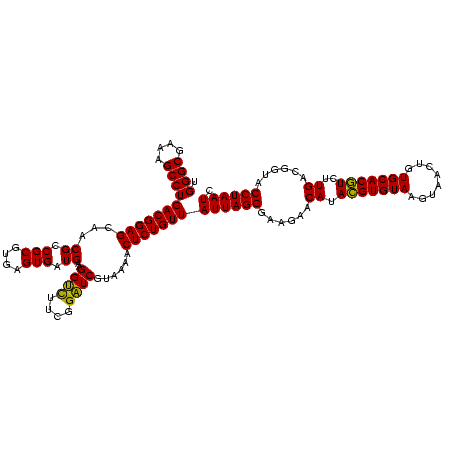
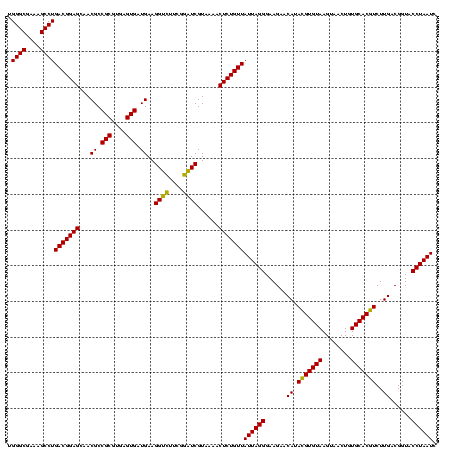
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -32.35 |
| Energy contribution | -32.13 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.659903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/760-880 CGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGA ((.(((..((.....))((((((((..(((.((....)).))).............((....((((((((.(.(((........)))..).))))))))..)).))))))))).)).)). ( -32.40) >sy_au.0 692085 120 - 2809422/760-880 CGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGA ((.(((..((.....))((((((((..(((.((....)).))).............((....((((((((.(.(((........)))..).))))))))..)).))))))))).)).)). ( -32.40) >sy_sa.0 841564 120 + 2516575/760-880 CGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGA ((.(((..((.....))((((((((..(((((....))..))).............((....((((((((.(.(((........)))..).))))))))..)).))))))))).)).)). ( -33.90) >consensus CGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGA ((.(((..((.....))((((((((..(((.((....)).))).............((....((((((((.(.(((........)))..).))))))))..)).))))))))).)).)). (-32.35 = -32.13 + -0.22)

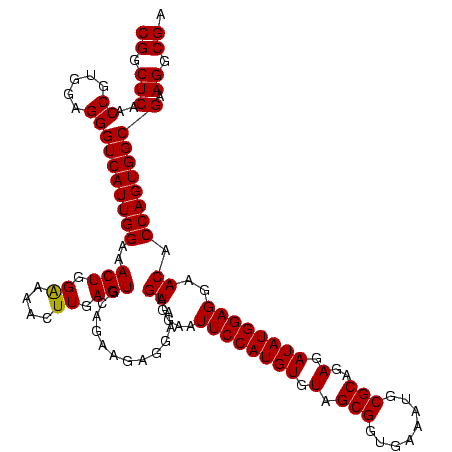
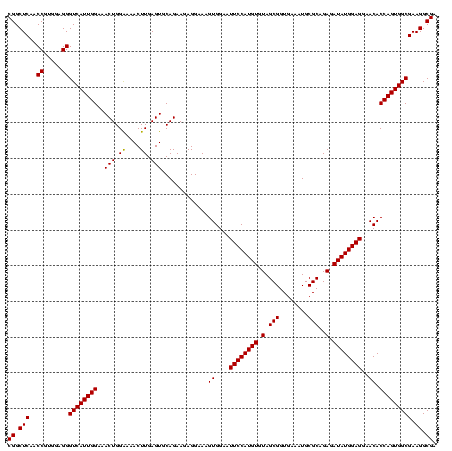
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -39.90 |
| Energy contribution | -39.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/800-920 AGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGG ((((((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))).))).(((((..........))))).. ( -39.90) >sy_au.0 692085 120 - 2809422/800-920 AGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGG ((((((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))).))).(((((..........))))).. ( -39.90) >sy_sa.0 841564 120 + 2516575/800-920 AGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGG ((((((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))).))).(((((..........))))).. ( -39.90) >consensus AGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGG ((((((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))).))).(((((..........))))).. (-39.90 = -39.90 + -0.00)

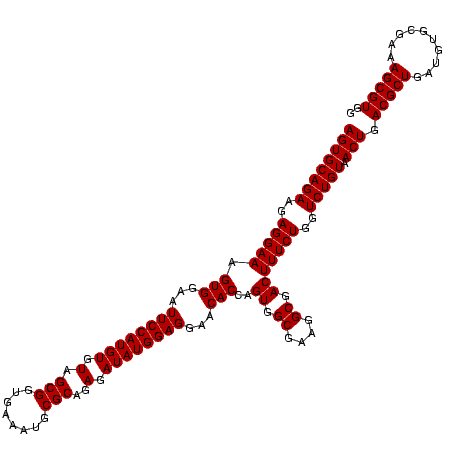
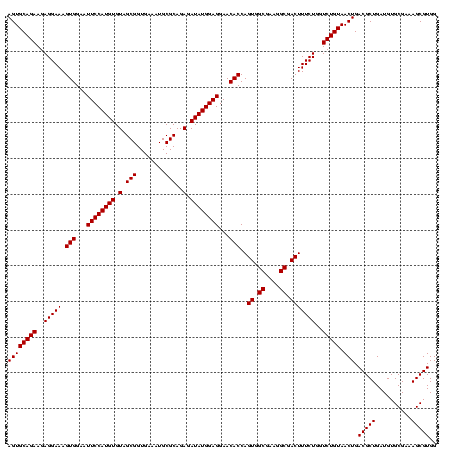
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -23.80 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/800-920 CCACGCUUUCGCACAUCAGCGUCAGUUACAGACCAGAAAGUCGCCUUCGCCACUGGUGUUCCUCCAUAUCUCUGCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACU ....(((..(((......)))..)))..((((...((((((((((.........))))....(((((......(((........))).....))))).....))))))....)))).... ( -23.80) >sy_au.0 692085 120 - 2809422/800-920 CCACGCUUUCGCACAUCAGCGUCAGUUACAGACCAGAAAGUCGCCUUCGCCACUGGUGUUCCUCCAUAUCUCUGCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACU ....(((..(((......)))..)))..((((...((((((((((.........))))....(((((......(((........))).....))))).....))))))....)))).... ( -23.80) >sy_sa.0 841564 120 + 2516575/800-920 CCACGCUUUCGCACAUCAGCGUCAGUUACAGACCAGAAAGUCGCCUUCGCCACUGGUGUUCCUCCAUAUCUCUGCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACU ....(((..(((......)))..)))..((((...((((((((((.........))))....(((((......(((........))).....))))).....))))))....)))).... ( -23.80) >consensus CCACGCUUUCGCACAUCAGCGUCAGUUACAGACCAGAAAGUCGCCUUCGCCACUGGUGUUCCUCCAUAUCUCUGCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACU ....(((..(((......)))..)))..((((...((((((((((.........))))....(((((......(((........))).....))))).....))))))....)))).... (-23.80 = -23.80 + 0.00) # Strand winner: forward (1.00)

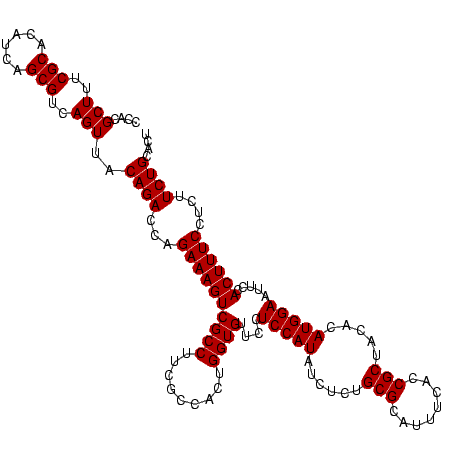
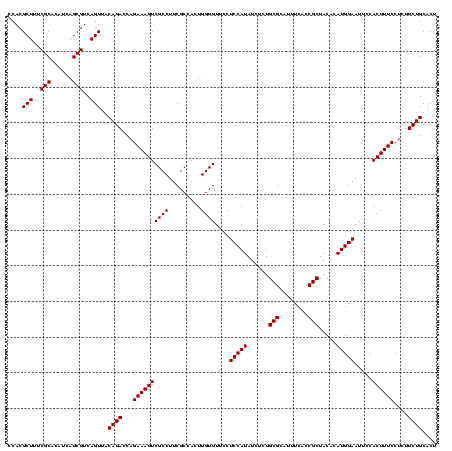
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -44.00 |
| Energy contribution | -46.00 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/920-1040 GGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUAC ((((....((((.........))))...))))((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).)).. ( -46.00) >sy_au.0 692085 120 - 2809422/920-1040 GGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUAC ((((....((((.........))))...))))((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).)).. ( -46.00) >sy_sa.0 841564 120 + 2516575/920-1040 GGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUAC ((((....((((.........))))...))))((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).)).. ( -46.00) >consensus GGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUAC ((((....((((.........))))...))))((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).)).. (-44.00 = -46.00 + 2.00)

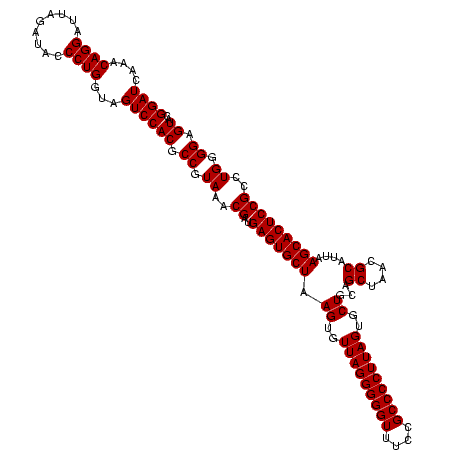
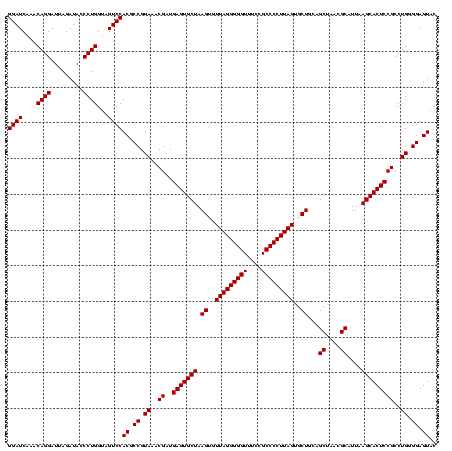
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -40.90 |
| Energy contribution | -40.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/920-1040 GUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGAUCC .......(((.((.(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..((....)))).))).....((((((.....))))))........ ( -40.90) >sy_au.0 692085 120 - 2809422/920-1040 GUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGAUCC .......(((.((.(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..((....)))).))).....((((((.....))))))........ ( -40.90) >sy_sa.0 841564 120 + 2516575/920-1040 GUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGAUCC .......(((.((.(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..((....)))).))).....((((((.....))))))........ ( -40.90) >consensus GUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGAUCC .......(((.((.(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..((....)))).))).....((((((.....))))))........ (-40.90 = -40.90 + -0.00) # Strand winner: forward (0.84)

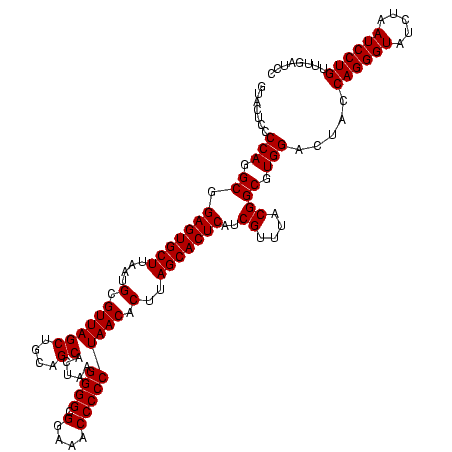
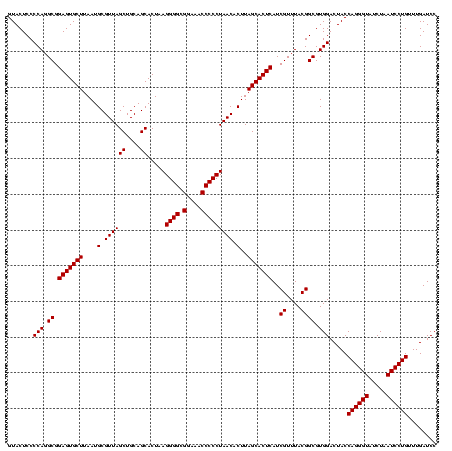
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -53.70 |
| Consensus MFE | -51.70 |
| Energy contribution | -53.70 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/960-1080 AACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCG ..(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))) ( -53.70) >sy_au.0 692085 120 - 2809422/960-1080 AACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCG ..(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))) ( -53.70) >sy_sa.0 841564 120 + 2516575/960-1080 AACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCG ..(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))) ( -53.70) >consensus AACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCG ..(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))) (-51.70 = -53.70 + 2.00)

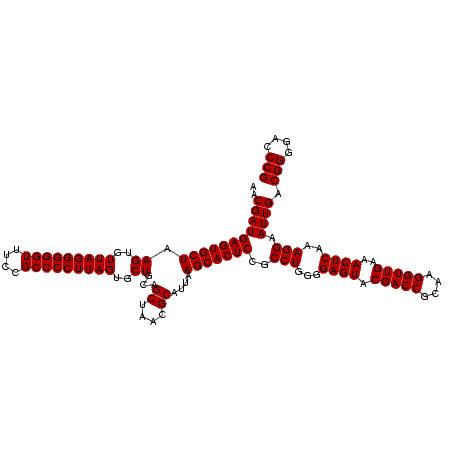
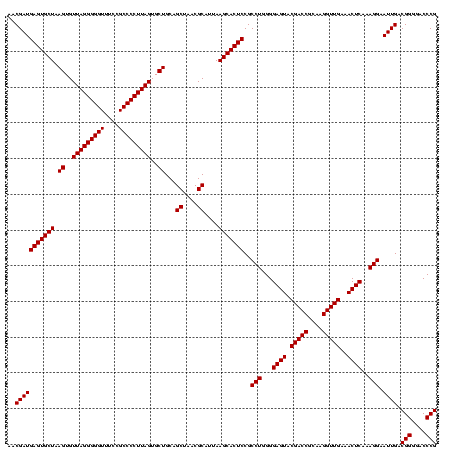
| Location | 971,068 – 971,188 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -40.30 |
| Energy contribution | -40.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 120 + 2685015/1240-1360 AAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGACUU ..((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))).. ( -40.90) >sy_au.0 692085 120 - 2809422/1240-1360 AAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGACUU ..((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))).. ( -40.90) >sy_sa.0 841564 120 + 2516575/1240-1360 AAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGACUU ..((((.((((((......))))))))))(((.......)))..(((.((((((.((......)).)))).....((((((..((((((.........))))))..)))))).))))).. ( -40.30) >consensus AAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGACUU ..((((.((((((......))))))))))(((.......)))..(((.((((((.((......)).)))).....((((((..((((((.........))))))..)))))).))))).. (-40.30 = -40.30 + 0.00) # Strand winner: reverse (0.84)

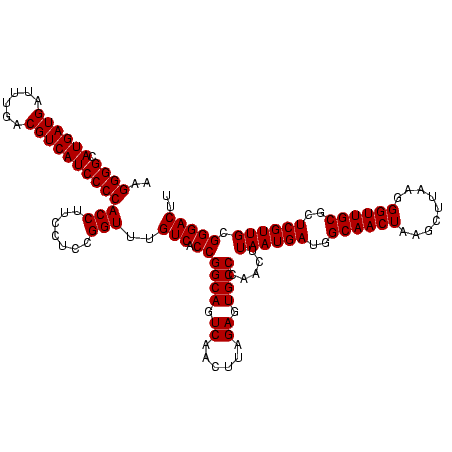
| Location | 971,068 – 971,186 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.37 |
| Consensus MFE | -43.80 |
| Energy contribution | -43.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 118 + 2685015/1560-1680 ACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUU--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGG .(((..((((((..(((((((...((((..(((..((....--)).))).)))))))..))))..))))))..)))...................((((((((((....)))))))))). ( -44.20) >sy_au.0 692085 119 - 2809422/1560-1680 ACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGG .(((..((((((..(((((((...((((..(((..((.....-)).))).)))))))..))))..))))))..)))...................((((((((((....)))))))))). ( -44.10) >sy_sa.0 841564 120 + 2516575/1560-1680 ACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGG .(((..((((((..(((((((...((((..(((..(((....))).))).)))))))..))))..))))))..)))...................((((((((((....)))))))))). ( -44.80) >consensus ACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUUA_GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGG .(((..((((((..(((((((...((((..(((..((......)).))).)))))))..))))..))))))..)))...................((((((((((....)))))))))). (-43.80 = -43.80 + 0.00)


| Location | 971,068 – 971,184 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -37.35 |
| Energy contribution | -37.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 116 + 2685015/1600-1720 U--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU-- .--.(((....(((.....(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))......))).....)))..-- ( -38.40) >sy_au.0 692085 119 - 2809422/1600-1720 UA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCCUC .(-((((.......((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))......))))). ( -40.02) >sy_sa.0 841564 120 + 2516575/1600-1720 UAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUA .((((......)))).(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((.....)))...)))))))... ( -40.60) >consensus UA_GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUU_ ...((((....(((.....(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))......))).....))))... (-37.35 = -37.47 + 0.11)


| Location | 971,068 – 971,183 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.18 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -34.83 |
| Energy contribution | -34.50 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 115 + 2685015/1640-1760 GAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---AGAGAUGA-GGAGUAACGU-GACAUAUUGUAUUCAGUUU .........((((..((((((((((....))))))))))..((((((((((((..((......))..)))((((((..---.)))))))-))))....))-))....))))......... ( -35.80) >sy_au.0 692085 118 - 2809422/1640-1760 GAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCCUCAGAAGAUGC-GGAAUAACGU-GACAUAUUGUAUUCAGUUU .........((((..((((((((((....))))))))))..((((.((((.....))))..(((((.(..(((((((....))))))))-.)))))..))-))....))))......... ( -37.90) >sy_sa.0 841564 120 + 2516575/1640-1760 GAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGGAAUAACAUUGACAUAUUGUAUUCAGUUU ...((((......((((((((((((....))))))))).....)))((((.....))))..(((((....(((((((....)))))))...)))))....))))................ ( -35.40) >consensus GAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUU__GAAGAUGC_GGAAUAACGU_GACAUAUUGUAUUCAGUUU ...(((.......((((((((((((....))))))))).....)))((((.....))))..(((((....(((((((....)))))))...))))).....)))................ (-34.83 = -34.50 + -0.33)

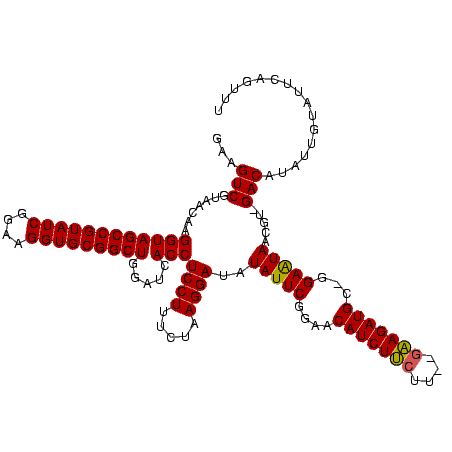
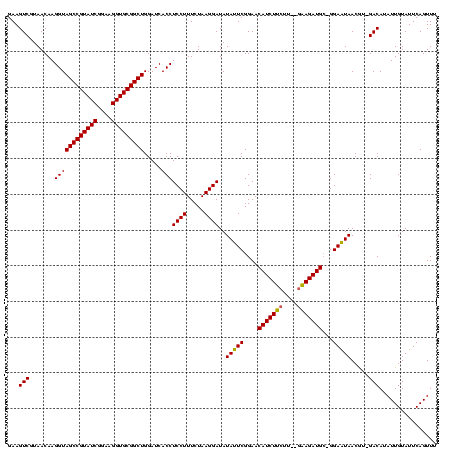
| Location | 971,068 – 971,179 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -28.08 |
| Energy contribution | -27.20 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 111 + 2685015/1668-1788 AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---AGAGAUGA-GGAGUAACGU-GACAUAUUGUAUUCAGUUUUGAAUGUUUAUUUUAAACAUUCA----A .(((((((.((..((((((((((((..((......))..)))((((((..---.)))))))-))))....))-))..))))))))).....(((((((((((...))))))))))----) ( -27.90) >sy_au.0 692085 112 - 2809422/1668-1788 AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCCUCAGAAGAUGC-GGAAUAACGU-GACAUAUUGUAUUCAGUUUUGAAUGUUUAUUU--AACAUUCA----A .(((((((.((..((((.((((.....))))..(((((.(..(((((((....))))))))-.)))))..))-))..))))))))).....(((((((((.....--))))))))----) ( -29.20) >sy_sa.0 841564 119 + 2516575/1668-1788 AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGGAAUAACAUUGACAUAUUGUAUUCAGUUUUGAAUGCUCAUUG-GAGUAUUCAUUGCA ...(((((((((((((..((((.....))))..(((((....(((((((....)))))))...)))))............)).)))))))).(((((((((....-)))))))))..))) ( -33.30) >consensus AGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUU__GAAGAUGC_GGAAUAACGU_GACAUAUUGUAUUCAGUUUUGAAUGUUUAUUU__AACAUUCA____A ......((((((((((..((((.....))))..(((((....(((((((....)))))))...)))))............)).)))))))).(((((((((.....)))))))))..... (-28.08 = -27.20 + -0.88)


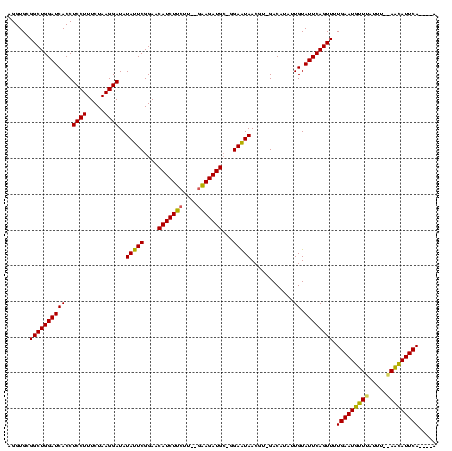
| Location | 971,068 – 971,179 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.55 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -24.67 |
| Energy contribution | -25.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974603 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 971068 111 + 2685015/1668-1788 U----UGAAUGUUUAAAAUAAACAUUCAAAACUGAAUACAAUAUGUC-ACGUUACUCC-UCAUCUCU---ACGAGAUGUUCCGAAUAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCU (----((((((((((...)))))))))))...............(((-((....((((-(....(((---(.(.(((((.......)))))).)))).))))))))))............ ( -23.40) >sy_au.0 692085 112 - 2809422/1668-1788 U----UGAAUGUU--AAAUAAACAUUCAAAACUGAAUACAAUAUGUC-ACGUUAUUCC-GCAUCUUCUGAGGAAGAUGUUCCGAAUAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCU (----((((((((--.....)))))))))...............(((-((..(((((.-((((((((....))))))))...)))))..((((.....)))).)))))............ ( -28.20) >sy_sa.0 841564 119 + 2516575/1668-1788 UGCAAUGAAUACUC-CAAUGAGCAUUCAAAACUGAAUACAAUAUGUCAAUGUUAUUCCUGCAUCUUCGUAAGAAGAUGUUCCGAAUAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCU (((..(((((.(((-....))).)))))...(((..(((.............(((((..((((((((....))))))))...)))))..((((.....)))).)))...)))..)))... ( -29.90) >consensus U____UGAAUGUU__AAAUAAACAUUCAAAACUGAAUACAAUAUGUC_ACGUUAUUCC_GCAUCUUC__AAGAAGAUGUUCCGAAUAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCU .....(((((((((.....)))))))))........................(((((..((((((((....))))))))...)))))..((((.....))))((((.........)))). (-24.67 = -25.23 + 0.56) # Strand winner: forward (0.99)

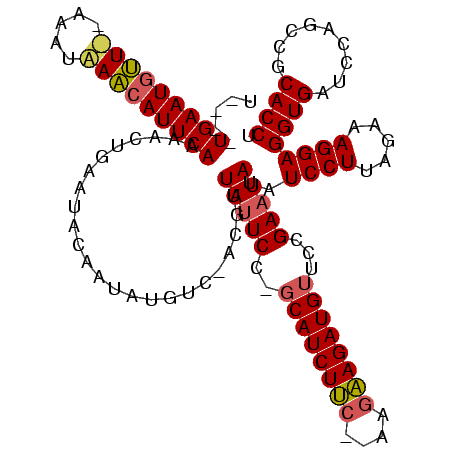
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:34 2006