
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 914,902 – 915,020 |
| Length | 118 |
| Max. P | 0.983200 |

| Location | 914,902 – 915,020 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -23.37 |
| Energy contribution | -24.70 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 914902 118 + 2685015/0-120 UUUUAAUUGACUUUUACUUAAAAAAUGAUAGAAUUUUAUUCGGGCUCCUUUAAAUAGGAC-ACGUGUAUAAAUCAUAUACGCUCCCCUUACAGA-UAUGUGAGAGGGAGCGAUUUUUUUA ...................(((((((....((.((.(((.((...((((......)))).-.)).))).)).)).....(((((((((((((..-..)))))).)))))))))))))).. ( -30.90) >sy_au.0 628144 118 - 2809422/0-120 UUUUGUUUGACUUUUAGUUGAAAGAUGAUAAAAUUUUAUUCGGGCUCCUUUAAAUAGGAC-ACGUGUAUAAAAUUUAUACGCUCCCCUUACAGAAUUUGUGAGAGGGAGCG-UUUUUUUA .....((..((.....))..))........(((((((((.((...((((......)))).-.))...)))))))))..(((((((((((((((...))))))).)))))))-)....... ( -34.30) >sy_sa.0 787904 120 + 2516575/0-120 AUUUAAUUGACUUUUUUCGUGAACAUGAUAAAAUUUUAUUCGGGCUCCUUUAAUUAGGACGACGUGUAUAAAUAUUAUACGCUCCCCCUUACAACCAUGUGAGAGGGAGCGAUUUUUUUG ...........((((.((((....)))).)))).(((((.((.((((((......)))).).).)).))))).......(((((((.((((((....)))))).)))))))......... ( -30.70) >consensus UUUUAAUUGACUUUUACUUAAAAAAUGAUAAAAUUUUAUUCGGGCUCCUUUAAAUAGGAC_ACGUGUAUAAAUAUUAUACGCUCCCCUUACAGA_UAUGUGAGAGGGAGCGAUUUUUUUA ..................................(((((.((...((((......))))...))...))))).......(((((((((((((.....)))))).)))))))......... (-23.37 = -24.70 + 1.33)

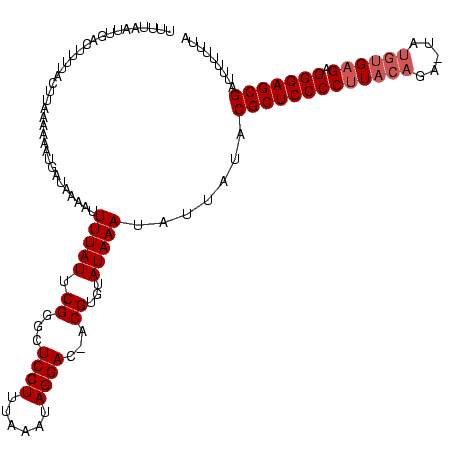
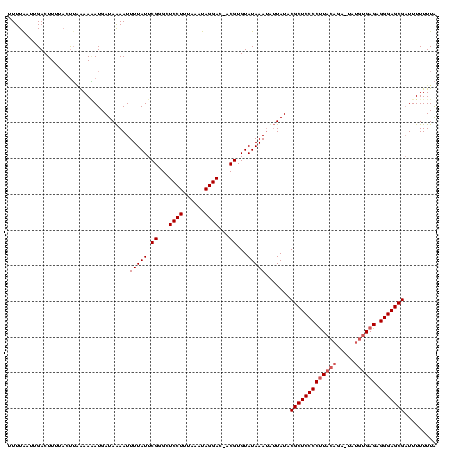
| Location | 914,902 – 915,020 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.12 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.54 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 914902 118 + 2685015/0-120 UAAAAAAAUCGCUCCCUCUCACAUA-UCUGUAAGGGGAGCGUAUAUGAUUUAUACACGU-GUCCUAUUUAAAGGAGCCCGAAUAAAAUUCUAUCAUUUUUUAAGUAAAAGUCAAUUAAAA .........(((((((.((.(((..-..))).)))))))))....((((((.(((.((.-(((((......)))).).))..(((((..........))))).))).))))))....... ( -27.60) >sy_au.0 628144 118 - 2809422/0-120 UAAAAAAA-CGCUCCCUCUCACAAAUUCUGUAAGGGGAGCGUAUAAAUUUUAUACACGU-GUCCUAUUUAAAGGAGCCCGAAUAAAAUUUUAUCAUCUUUCAACUAAAAGUCAAACAAAA .......(-(((((((.((.(((.....))).))))))))))..(((((((((...((.-(((((......)))).).)).))))))))).............................. ( -26.30) >sy_sa.0 787904 120 + 2516575/0-120 CAAAAAAAUCGCUCCCUCUCACAUGGUUGUAAGGGGGAGCGUAUAAUAUUUAUACACGUCGUCCUAAUUAAAGGAGCCCGAAUAAAAUUUUAUCAUGUUCACGAAAAAAGUCAAUUAAAU ..........(((((((((.(((....))).)))))))))(((((.....)))))...(((((((......))))....(((((...........))))).)))................ ( -29.50) >consensus UAAAAAAAUCGCUCCCUCUCACAUA_UCUGUAAGGGGAGCGUAUAAAAUUUAUACACGU_GUCCUAUUUAAAGGAGCCCGAAUAAAAUUUUAUCAUCUUUAAAAUAAAAGUCAAUUAAAA ..........((((((.((.(((.....))).))))))))(((((.....))))).((...((((......))))...))........................................ (-20.22 = -21.00 + 0.78) # Strand winner: forward (1.00)

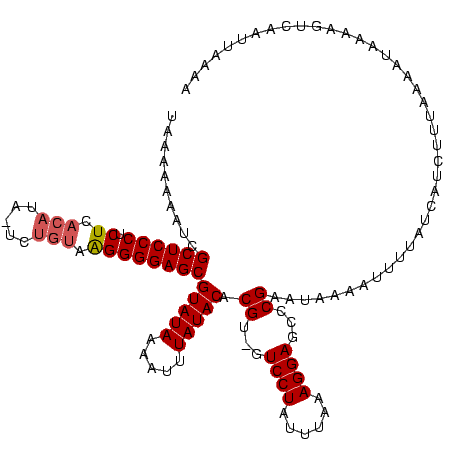
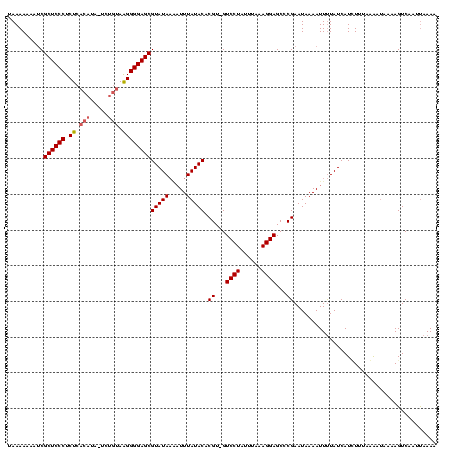
| Location | 914,902 – 915,020 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -25.74 |
| Energy contribution | -26.63 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.42 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 914902 118 + 2685015/3-123 UAAUUGACUUUUACUUAAAAAAUGAUAGAAUUUUAUUCGGGCUCCUUUAAAUAGGAC-ACGUGUAUAAAUCAUAUACGCUCCCCUUACAGA-UAUGUGAGAGGGAGCGAUUUUUUUAUUU ...................((((((.((((((.......(..((((......)))).-.)((((((.....))))))((((((((((((..-..)))))).)))))))))))).)))))) ( -31.60) >sy_au.0 628144 118 - 2809422/3-123 UGUUUGACUUUUAGUUGAAAGAUGAUAAAAUUUUAUUCGGGCUCCUUUAAAUAGGAC-ACGUGUAUAAAAUUUAUACGCUCCCCUUACAGAAUUUGUGAGAGGGAGCG-UUUUUUUAUUU ..((..((.....))..))((((((..(((((((((.((...((((......)))).-.))...)))))))))..(((((((((((((((...))))))).)))))))-)....)))))) ( -35.10) >sy_sa.0 787904 120 + 2516575/3-123 UAAUUGACUUUUUUCGUGAACAUGAUAAAAUUUUAUUCGGGCUCCUUUAAUUAGGACGACGUGUAUAAAUAUUAUACGCUCCCCCUUACAACCAUGUGAGAGGGAGCGAUUUUUUUGUUU .................(((((..((((.(((((((.((...((((......))))...)).))).)))).)))).(((((((.((((((....)))))).))))))).......))))) ( -31.30) >consensus UAAUUGACUUUUACUUAAAAAAUGAUAAAAUUUUAUUCGGGCUCCUUUAAAUAGGAC_ACGUGUAUAAAUAUUAUACGCUCCCCUUACAGA_UAUGUGAGAGGGAGCGAUUUUUUUAUUU .......................(((((((.(((((.((...((((......))))...))...))))).......(((((((((((((.....)))))).)))))))....))))))). (-25.74 = -26.63 + 0.89)

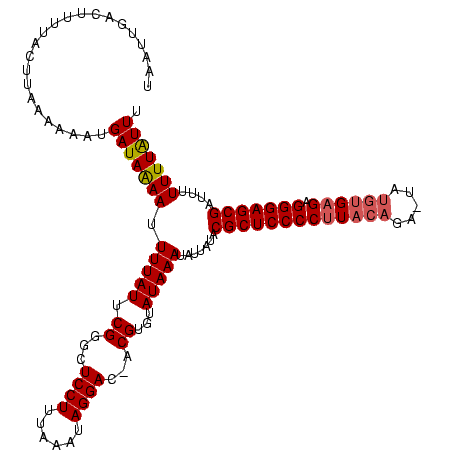
| Location | 914,902 – 915,020 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.68 |
| Mean single sequence MFE | -27.80 |
| Consensus MFE | -20.22 |
| Energy contribution | -21.00 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 914902 118 + 2685015/3-123 AAAUAAAAAAAUCGCUCCCUCUCACAUA-UCUGUAAGGGGAGCGUAUAUGAUUUAUACACGU-GUCCUAUUUAAAGGAGCCCGAAUAAAAUUCUAUCAUUUUUUAAGUAAAAGUCAAUUA ............(((((((.((.(((..-..))).)))))))))....((((((.(((.((.-(((((......)))).).))..(((((..........))))).))).)))))).... ( -27.60) >sy_au.0 628144 118 - 2809422/3-123 AAAUAAAAAAA-CGCUCCCUCUCACAAAUUCUGUAAGGGGAGCGUAUAAAUUUUAUACACGU-GUCCUAUUUAAAGGAGCCCGAAUAAAAUUUUAUCAUCUUUCAACUAAAAGUCAAACA ..........(-(((((((.((.(((.....))).))))))))))..(((((((((...((.-(((((......)))).).)).)))))))))........................... ( -26.30) >sy_sa.0 787904 120 + 2516575/3-123 AAACAAAAAAAUCGCUCCCUCUCACAUGGUUGUAAGGGGGAGCGUAUAAUAUUUAUACACGUCGUCCUAAUUAAAGGAGCCCGAAUAAAAUUUUAUCAUGUUCACGAAAAAAGUCAAUUA .............(((((((((.(((....))).)))))))))(((((.....)))))...(((((((......))))....(((((...........))))).)))............. ( -29.50) >consensus AAAUAAAAAAAUCGCUCCCUCUCACAUA_UCUGUAAGGGGAGCGUAUAAAAUUUAUACACGU_GUCCUAUUUAAAGGAGCCCGAAUAAAAUUUUAUCAUCUUUAAAAUAAAAGUCAAUUA .............((((((.((.(((.....))).))))))))(((((.....))))).((...((((......))))...))..................................... (-20.22 = -21.00 + 0.78) # Strand winner: forward (1.00)

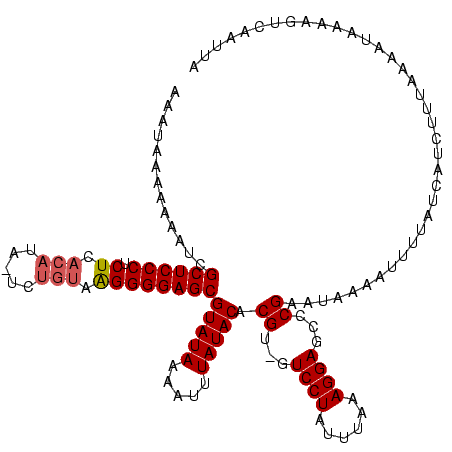

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:13 2006