
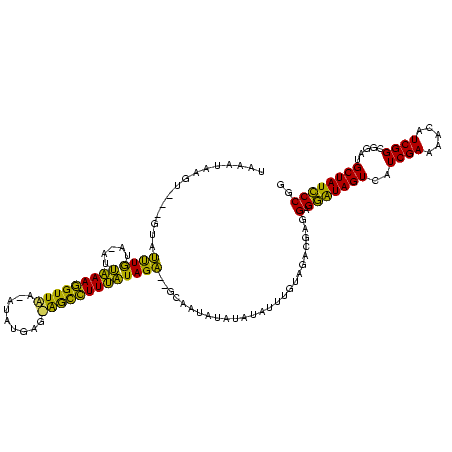
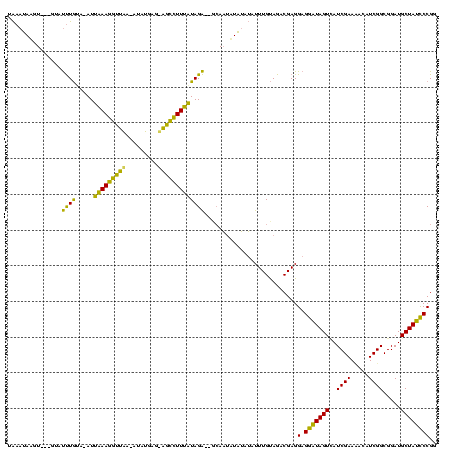
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 888,823 – 888,943 |
| Length | 120 |
| Max. P | 0.995212 |

| Location | 888,823 – 888,934 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.73 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -16.92 |
| Energy contribution | -13.50 |
| Covariance contribution | -3.42 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581632 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 888823 111 + 2685015/160-280 -AUAUAAGU---GUACCUAUA-AUCAAAAGUUAA-AUAUGAG-AGCUUUUGUAGG--GCAAUUUAUUUCUUUGCAGACGAGGAGAAUAGUCAUCGAAAAUAUCGGCGGAUGCUAUUCCGG -..((((((---...((((..-..((((((((..-.......-))))))))))))--...)))))).((((((....))))))((((((.((((.............))))))))))... ( -24.02) >sy_au.0 600745 117 - 2809422/160-280 UAAAUAUUU---GUAUUUGAAGAUUAAAGGUUAAUAUAUGAGUGGCCUUUAUAGAGUGCAAUAUAUGUAUUUGUAGACGAGGAGGAUAGUGAUCGAAUAGAUCGGCGGAUGCUAUCCCGG (((((((((---((((((......(((((((((.........)))))))))..)))))))).....)))))))........(.(((((((((((.....)))).......)))))))).. ( -25.31) >sy_sa.0 759250 116 + 2516575/160-280 UGAAGAGGUUAUGUGUUUGUA-AUUUAAGGUUUA--UAACAGAAAUCUUAAUAGACACGAUAAUCGAAGUUUGUAGACGAGGAGGGUAGUUAUCGAAC-CAUCGGCGGAUGCUAUCCCGG ......((((((((((((((.-...((((((((.--......)))))))))))))))).))))))............((.(((.((((.(..((((..-..))))..).)))).))))). ( -29.20) >consensus UAAAUAAGU___GUAUUUGUA_AUUAAAGGUUAA_AUAUGAG_AGCCUUUAUAGA__GCAAUAUAUAUAUUUGUAGACGAGGAGGAUAGUCAUCGAAAACAUCGGCGGAUGCUAUCCCGG ...............((((.....(((((((((.........)))))))))))))..........................(.(((((((..((((.....)))).....)))))))).. (-16.92 = -13.50 + -3.42)

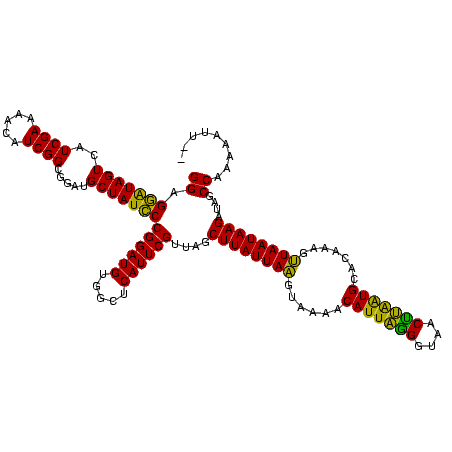
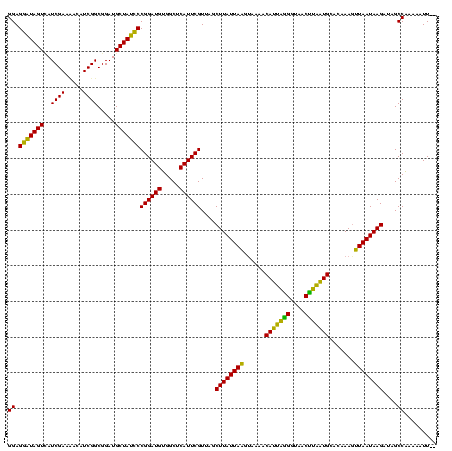
| Location | 888,823 – 888,943 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -29.92 |
| Energy contribution | -28.49 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 888823 120 + 2685015/240-360 GGAGAAUAGUCAUCGAAAAUAUCGGCGGAUGCUAUUCCGGAUGAGGCUCAUUCGUUAGCUUAUUAUUUAAAACAUUUAGGUAACUAAAUGCACAAAAAUAAUAAGAAUGCCUAAAAUUUA ((.((((((.((((.............))))))))))((((((.....))))))....((((((((((....(((((((....))))))).....))))))))))....))......... ( -28.82) >sy_au.0 600745 118 - 2809422/240-360 GGAGGAUAGUGAUCGAAUAGAUCGGCGGAUGCUAUCCCGGAUGUGGCUCAUUCGUUAGCUUAUUAAGUAAAACAUUAGGGUGACUUAAUGGACAAAGUUAAUAAGAUCGCCAGAAAUU-- ..........((((.....))))(((((..((((...((((((.....)))))).))))(((((((.(....(((((((....))))))).....).)))))))..))))).......-- ( -35.00) >sy_sa.0 759250 117 + 2516575/240-360 GGAGGGUAGUUAUCGAAC-CAUCGGCGGAUGCUAUCCCGGAUGUGGCUCAUUCGUUAACUUAUUAAGGAAAACAUUAGGGUAACCUGGUGCACAAAGUUAAUAAGAUAGCCAAAAAUU-- (((.((((.(..((((..-..))))..).)))).))).(((((.....)))))((((.((((((((......(((((((....))))))).......)))))))).))))........-- ( -30.02) >consensus GGAGGAUAGUCAUCGAAAACAUCGGCGGAUGCUAUCCCGGAUGUGGCUCAUUCGUUAGCUUAUUAAGUAAAACAUUAGGGUAACUUAAUGCACAAAGUUAAUAAGAUAGCCAAAAAUU__ ((.(((((((..((((.....)))).....)))))))((((((.....))))))....((((((((......(((((((....))))))).......))))))))....))......... (-29.92 = -28.49 + -1.43)

| Location | 888,823 – 888,943 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -20.11 |
| Energy contribution | -19.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 888823 120 + 2685015/240-360 UAAAUUUUAGGCAUUCUUAUUAUUUUUGUGCAUUUAGUUACCUAAAUGUUUUAAAUAAUAAGCUAACGAAUGAGCCUCAUCCGGAAUAGCAUCCGCCGAUAUUUUCGAUGACUAUUCUCC ........((((...((((((((((....((((((((....))))))))...))))))))))(....).....)))).....(((((((((((....((.....)))))).))))))).. ( -28.90) >sy_au.0 600745 118 - 2809422/240-360 --AAUUUCUGGCGAUCUUAUUAACUUUGUCCAUUAAGUCACCCUAAUGUUUUACUUAAUAAGCUAACGAAUGAGCCACAUCCGGGAUAGCAUCCGCCGAUCUAUUCGAUCACUAUCCUCC --.......((((...(((((((....(..(((((........)))))..)...)))))))((((.((.(((.....))).))...))))...))))((((.....)))).......... ( -21.60) >sy_sa.0 759250 117 + 2516575/240-360 --AAUUUUUGGCUAUCUUAUUAACUUUGUGCACCAGGUUACCCUAAUGUUUUCCUUAAUAAGUUAACGAAUGAGCCACAUCCGGGAUAGCAUCCGCCGAUG-GUUCGAUAACUACCCUCC --......(((((...(..(((((((...(((..(((....)))..)))..........)))))))..)...))))).....(((.(((.(((.(((...)-))..)))..))))))... ( -22.22) >consensus __AAUUUUUGGCAAUCUUAUUAACUUUGUGCAUUAAGUUACCCUAAUGUUUUACUUAAUAAGCUAACGAAUGAGCCACAUCCGGGAUAGCAUCCGCCGAUAUAUUCGAUAACUAUCCUCC ........((((...((((((((......((((((((....)))))))).....))))))))(....).....)))).....(((((((.......(((.....)))....))))))).. (-20.11 = -19.57 + -0.55) # Strand winner: forward (1.00)

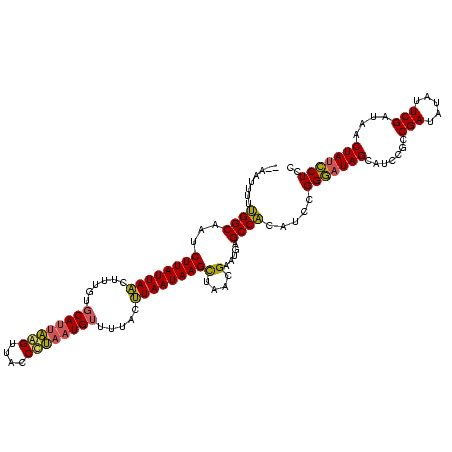
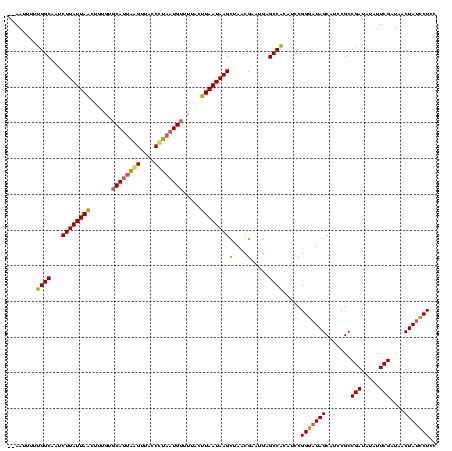
| Location | 888,823 – 888,943 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -27.62 |
| Consensus MFE | -25.64 |
| Energy contribution | -24.65 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 888823 120 + 2685015/253-373 CGAAAAUAUCGGCGGAUGCUAUUCCGGAUGAGGCUCAUUCGUUAGCUUAUUAUUUAAAACAUUUAGGUAACUAAAUGCACAAAAAUAAUAAGAAUGCCUAAAAUUUAUAAAAUCUUAGAA ..........((((...((((...((((((.....)))))).))))(((((((((....(((((((....))))))).....)))))))))...))))...................... ( -26.20) >sy_au.0 600745 116 - 2809422/253-373 CGAAUAGAUCGGCGGAUGCUAUCCCGGAUGUGGCUCAUUCGUUAGCUUAUUAAGUAAAACAUUAGGGUGACUUAAUGGACAAAGUUAAUAAGAUCGCCAGAAAUU----GAAUAUAAAAA ..........(((((..((((...((((((.....)))))).))))(((((((.(....(((((((....))))))).....).)))))))..))))).......----........... ( -30.80) >sy_sa.0 759250 115 + 2516575/253-373 CGAAC-CAUCGGCGGAUGCUAUCCCGGAUGUGGCUCAUUCGUUAACUUAUUAAGGAAAACAUUAGGGUAACCUGGUGCACAAAGUUAAUAAGAUAGCCAAAAAUU----AAAUAUAAAAA .....-((((.(.((((...))))).))))(((((..((..(((((((...........(((((((....)))))))....)))))))..))..)))))......----........... ( -25.86) >consensus CGAAAACAUCGGCGGAUGCUAUCCCGGAUGUGGCUCAUUCGUUAGCUUAUUAAGUAAAACAUUAGGGUAACUUAAUGCACAAAGUUAAUAAGAUAGCCAAAAAUU____AAAUAUAAAAA ..........(((((((...))))((((((.....))))))....((((((((......(((((((....))))))).......))))))))...)))...................... (-25.64 = -24.65 + -0.99)

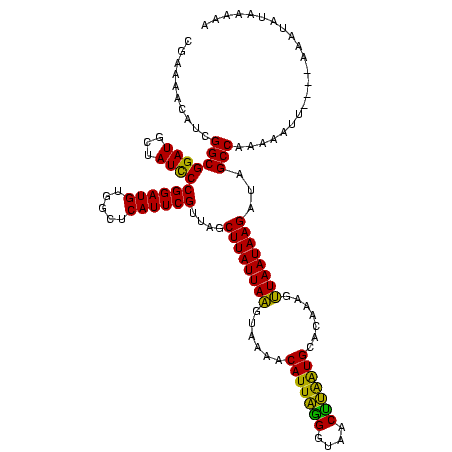
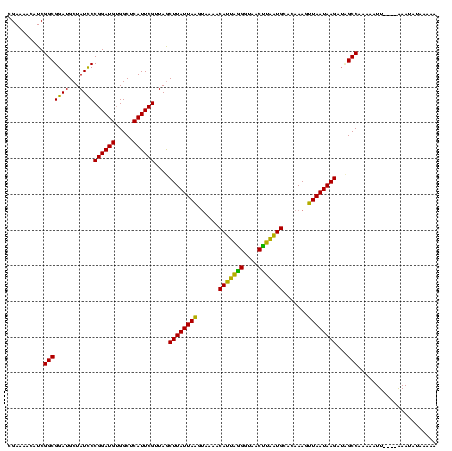
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:09 2006