| Sequence ID | sy_ha.0 |
|---|---|
| Location | 833,342 – 833,459 |
| Length | 117 |
| Max. P | 0.988606 |

| Location | 833,342 – 833,459 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.62 |
| Mean single sequence MFE | -19.90 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 833342 117 + 2685015/0-120 UAAAUACACUAACUACUUAUAUGUAAAGCAAAUGAGUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUGCCCCAUCAA--UUAAAUAUAU-UUCAAAGCGUGAACU ......(((....(((......)))..(((..(((((.....(((((((((((.......)))))))))))..)))))...)))........--..........-........))).... ( -22.40) >sy_au.0 515861 118 - 2809422/0-120 UAAAUACACUAACUACUUUUAUAUAAAGCAAAUGAGUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUACCCCAUCAA--CUAAAUAAAGAUUUAAAGCGUGAACU ......(((..(((.((((.....))))....(((((.....(((((((((((.......)))))))))))..))))).)))..........--.(((((....)))))....))).... ( -17.90) >sy_sa.0 712595 119 + 2516575/0-120 UAAAUACAUGUACUACU-AUAUAUAAAGCAAAUGAGUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUACCCCAUCAAAACUAAAUAAAACUUUAAAGCGUGAACU ......(((((......-..............(((((.....(((((((((((.......)))))))))))..)))))((........)).....................))))).... ( -19.40) >consensus UAAAUACACUAACUACUUAUAUAUAAAGCAAAUGAGUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUACCCCAUCAA__CUAAAUAAAA_UUUAAAGCGUGAACU ...........................((...(((((.....(((((((((((.......)))))))))))..)))))((........)).....................))....... (-17.10 = -17.10 + -0.00)


| Location | 833,342 – 833,459 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.90 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -34.40 |
| Energy contribution | -33.96 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -4.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
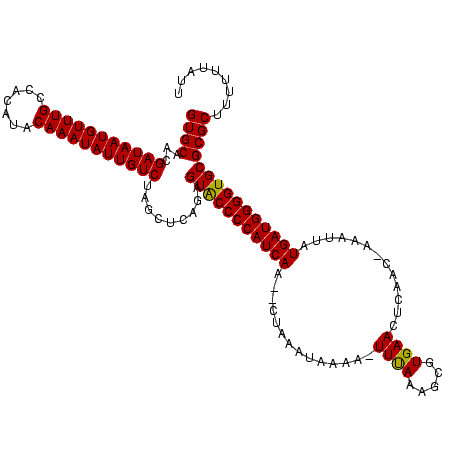
>sy_ha.0 833342 117 + 2685015/35-155 GUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUGCCCCAUCAA--UUAAAUAUAU-UUCAAAGCGUGAACUCAACUAAAAUAUGAUGGGGUGCGCGCUUUUUUAUU ((((...(((((((((((.......))))))))))).........(..((((((((.--..........-((((.....)))).............))))))))..)))))......... ( -34.35) >sy_au.0 515861 118 - 2809422/35-155 GUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUACCCCAUCAA--CUAAAUAAAGAUUUAAAGCGUGAACUCAACAAACUUAUGAUGGGGUGCGCGCUUUUUUAUU ((((...(((((((((((.......))))))))))).........(((((((((((.--.(((((....)))))(((..((.......))..))).)))))))))))))))......... ( -36.70) >sy_sa.0 712595 118 + 2516575/35-155 GUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUACCCCAUCAAAACUAAAUAAAACUUUAAAGCGUGAACUCAA--AAAUUAUGAUGGGGUGCGCGCUUUUUUAUA ((((...(((((((((((.......))))))))))).........(((((((((((....((((......))))..(.(....).)..--......)))))))))))))))......... ( -35.00) >consensus GUGCAACGAUAAUGUUUGCCACAUACAAAUAUUGUCUAGCUCAGAGUACCCCAUCAA__CUAAAUAAAA_UUUAAAGCGUGAACUCAAC_AAAUUAUGAUGGGGUGCGCGCUUUUUUAUU ((((...(((((((((((.......))))))))))).........(((((((((((..............((((.....)))).............)))))))))))))))......... (-34.40 = -33.96 + -0.44)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:59 2006