| Sequence ID | sp_pn.0 |
|---|---|
| Location | 1,789,618 – 1,789,724 |
| Length | 106 |
| Max. P | 0.999597 |

| Location | 1,789,618 – 1,789,722 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -33.53 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
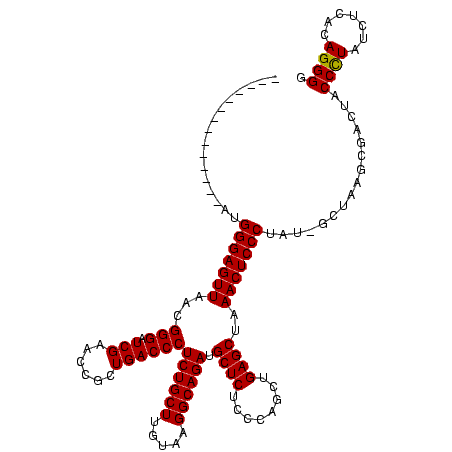
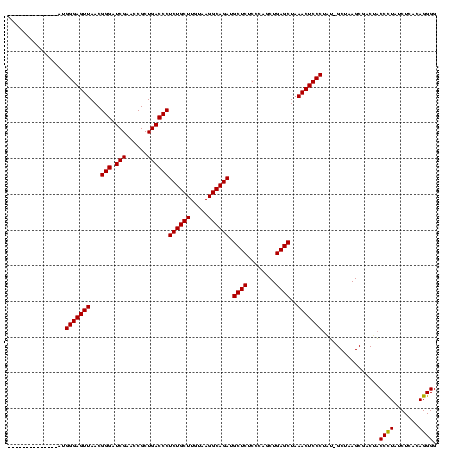
>sp_pn.0 1789618 104 + 2038615/40-160 CGAGUCUAAAAUAACUUGCGUUAUCUUAAA---CGGUCCCGACGGGAAUCGAACCCGCGAUCUUCGCCGUGACAGGGCGACGUGAUAACCGCUACACUACGGGACCU------------- ...((.(((.(((((....))))).))).)---)(((((((..(((.......)))((((((.(((((.......)))))...)))...))).......))))))).------------- ( -35.00) >sp_ag.0 2138742 82 - 2160267/40-160 CGAGUCUA-------------------------CGGUCCCGACGGGAAUCGAACCCGCGAUCUUCGCCGUGACAGGGCGACGUGAUAACCGCUACACUACGGGACCU------------- ........-------------------------.(((((((..(((.......)))((((((.(((((.......)))))...)))...))).......))))))).------------- ( -30.80) >sp_mu.0 1683671 120 + 2030921/40-160 CGAGUCGAUGAUCCCUCUCUUUUCAGAAAAAGACGGUCCCGACGGGAAUCGAACCCGCGAUCUUCGCCGUGACAGGGCGACGUGAUAACCGCUACACCACGGGACCUCUAACUAAUCACU .(((..(......)..)))...........(((.(((((((..(((.......)))((((((.(((((.......)))))...)))...))).......))))))))))........... ( -34.80) >consensus CGAGUCUA__AU__CU__C_UU_____AAA___CGGUCCCGACGGGAAUCGAACCCGCGAUCUUCGCCGUGACAGGGCGACGUGAUAACCGCUACACUACGGGACCU_____________ ..................................(((((((..(((.......)))((((((.(((((.......)))))...)))...))).......))))))).............. (-30.80 = -30.80 + 0.00)


| Location | 1,789,618 – 1,789,722 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.26 |
| Mean single sequence MFE | -41.53 |
| Consensus MFE | -32.50 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999597 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1789618 104 + 2038615/40-160 -------------AGGUCCCGUAGUGUAGCGGUUAUCACGUCGCCCUGUCACGGCGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCG---UUUAAGAUAACGCAAGUUAUUUUAGACUCG -------------.(((((((.......((((((......(((((.......))))).))))))(((.......)))..)))))))(---(((((((((((....))))))))))))... ( -47.30) >sp_ag.0 2138742 82 - 2160267/40-160 -------------AGGUCCCGUAGUGUAGCGGUUAUCACGUCGCCCUGUCACGGCGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCG-------------------------UAGACUCG -------------.(((((((.......((((((......(((((.......))))).))))))(((.......)))..))))))).-------------------------........ ( -32.50) >sp_mu.0 1683671 120 + 2030921/40-160 AGUGAUUAGUUAGAGGUCCCGUGGUGUAGCGGUUAUCACGUCGCCCUGUCACGGCGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCGUCUUUUUCUGAAAAGAGAGGGAUCAUCGACUCG .......((((.((((((((((((((.......)))))).(((((.......))))).....(((((.......))))).))))))((((((((((....))))))))))..)))))).. ( -44.80) >consensus _____________AGGUCCCGUAGUGUAGCGGUUAUCACGUCGCCCUGUCACGGCGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCG___UUU_____AA_G__AG__AU__UAGACUCG ..............(((((((.......((((((......(((((.......))))).))))))(((.......)))..))))))).................................. (-32.50 = -32.50 + 0.00) # Strand winner: reverse (0.92)

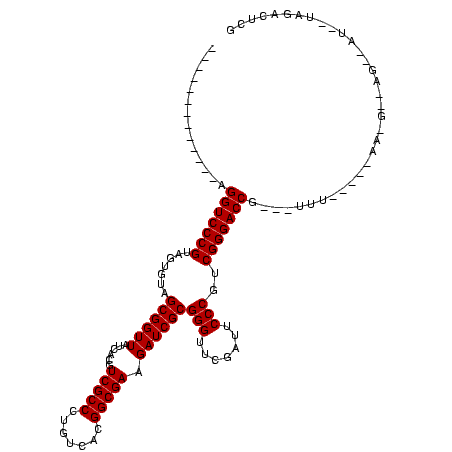
| Location | 1,789,618 – 1,789,724 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -33.19 |
| Energy contribution | -32.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1789618 106 + 2038615/160-280 --------------UUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUAGAGCUAAGCGACUUCCCUAUCUCACAGGGG --------------.((((((.....(((.(((......))))))((((((.....))))))....))))))(((.((((...........)))).))).....((((.......)))). ( -34.30) >sp_ag.0 2138742 105 - 2160267/160-280 --------------AUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUUU-GCUAAGCGACUACCUUAUCUCACAGGGG --------------..(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).((-((...))))...((((.......)))). ( -31.90) >sp_mu.0 1683671 120 + 2030921/160-280 UUAGUUACUUAGUUAUGGGAGUUAACGGGCUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAACUGAGCUAAACUCCCUCUUGCUAAGCGACGACCCUAUCUCACAGGGG ...((..((((((...(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))....))))))..))..((((.......)))). ( -44.70) >consensus ______________AUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUAU_GCUAAGCGACUACCCUAUCUCACAGGGG ................(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))................((((.......)))). (-33.19 = -32.97 + -0.22)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:50:17 2006