| Sequence ID | sy_ha.0 |
|---|---|
| Location | 765,300 – 1,588,702 |
| Length | 823402 |
| Max. P | 0.999678 |

| Location | 1,588,593 – 1,588,702 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.77 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.24 |
| Mean z-score | -6.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1588593 109 - 2685015 GGCAAGGUUGACUAGAAUU-AAAAAAGAUUGUUACAAGUGCAUUUUUA-GCCAGUCAACUACUGCCAAUAUAUAUUUG--AAGCCUGAGGCAUUGAAUUAUGCCACAGGCUCA ((((.((((((((.(...(-(((((.((((......))).).))))))-.).))))))))..))))..........((--(.(((((.(((((......))))).)))))))) ( -36.10) >sy_au.0 808671 112 + 2809422 GGCAAGGUUGACUAGAAUUGAAAAAAGCUUGUUACAAGCGCAUUUUCG-UUCAGUCAACUACUGCCAAUAUAACUUUGUAGAGCAUUGAACAUUGAUUUAUGUCUCAAGCUCA ((((.((((((((.((((.(((((..(((((...)))))...))))))-)))))))))))..))))..............((((.((((((((......)))).)))))))). ( -38.80) >sy_sa.0 622241 110 + 2516575 GGCAAGGAUGACUAGGAUUGAAAAUAGCUUGAUAUAAGCGCAUUUUCACUUCAGUCAUCUACUGCCUAAAUGAAAAAG---GGUCUGAGACAUCUAUUUUUGUCUCAGACUCU ((((.((((((((.(((.(((((((.(((((...)))))..))))))).)))))))))))..))))..........((---(((((((((((........))))))))))))) ( -49.20) >consensus GGCAAGGUUGACUAGAAUUGAAAAAAGCUUGUUACAAGCGCAUUUUCA_UUCAGUCAACUACUGCCAAUAUAAAUUUG___AGCCUGAGACAUUGAUUUAUGUCUCAGGCUCA ((((.((((((((.(((.((((((..(((((...)))))...)))))).)))))))))))..))))...............((((((((((((......)))))))))))).. (-36.20 = -36.77 + 0.57)

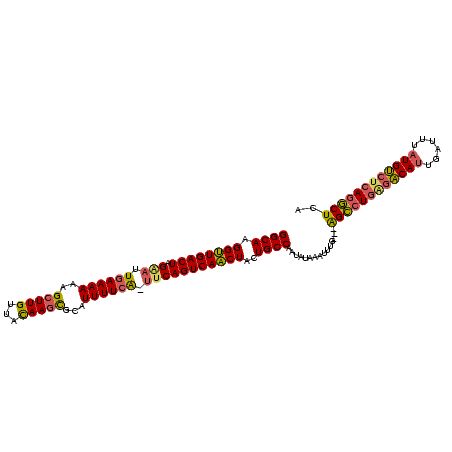

| Location | 1,588,593 – 1,588,702 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -29.10 |
| Energy contribution | -31.00 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.16 |
| Mean z-score | -5.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
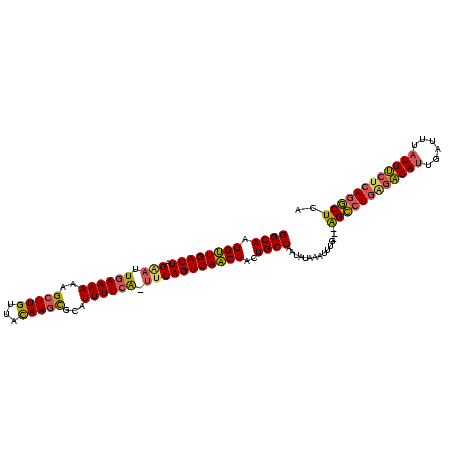
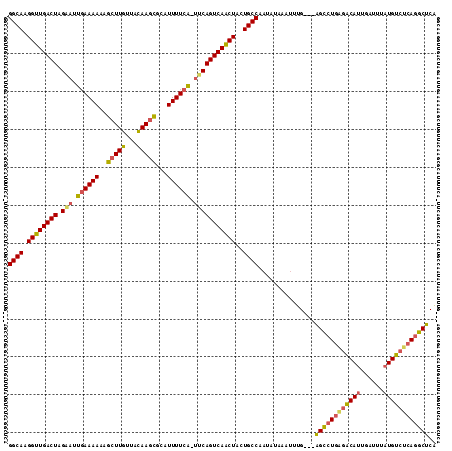
>sy_ha.0 1588593 109 - 2685015 UGAGCCUGUGGCAUAAUUCAAUGCCUCAGGCUU--CAAAUAUAUAUUGGCAGUAGUUGACUGGC-UAAAAAUGCACUUGUAACAAUCUUUUUU-AAUUCUAGUCAACCUUGCC .(((((((.(((((......))))).)))))))--............(((((..(((((((((.-(((((((((....))).......)))))-)...))))))))).))))) ( -36.11) >sy_au.0 808671 112 + 2809422 UGAGCUUGAGACAUAAAUCAAUGUUCAAUGCUCUACAAAGUUAUAUUGGCAGUAGUUGACUGAA-CGAAAAUGCGCUUGUAACAAGCUUUUUUCAAUUCUAGUCAACCUUGCC .(((((((((.(((......)))))))).))))..............(((((..((((((((((-.(((((...(((((...)))))..)))))..))).))))))).))))) ( -32.50) >sy_sa.0 622241 110 + 2516575 AGAGUCUGAGACAAAAAUAGAUGUCUCAGACC---CUUUUUCAUUUAGGCAGUAGAUGACUGAAGUGAAAAUGCGCUUAUAUCAAGCUAUUUUCAAUCCUAGUCAUCCUUGCC ((.((((((((((........)))))))))).---))..........(((((..((((((((...((((((((.((((.....))))))))))))....)))))))).))))) ( -41.40) >consensus UGAGCCUGAGACAUAAAUCAAUGUCUCAGGCU___CAAAUUUAUAUUGGCAGUAGUUGACUGAA_UGAAAAUGCGCUUGUAACAAGCUUUUUUCAAUUCUAGUCAACCUUGCC .(((((((.(((((......))))).)))))))..............(((((..((((((((((.((((((...((((.....))))..)))))).))).))))))).))))) (-29.10 = -31.00 + 1.90) # Strand winner: forward (1.00)

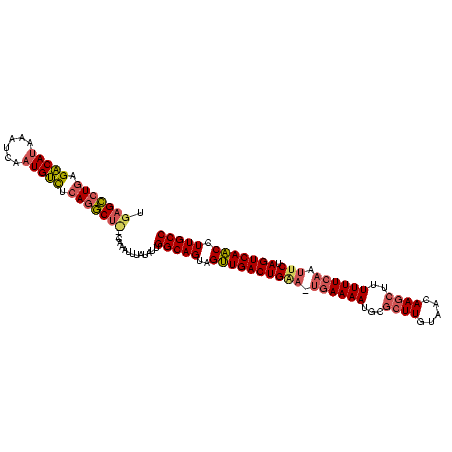
| Location | 1,588,593 – 1,588,702 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -41.37 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.77 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.24 |
| Mean z-score | -6.24 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1588593 109 - 2685015/0-113 GGCAAGGUUGACUAGAAUU-AAAAAAGAUUGUUACAAGUGCAUUUUUA-GCCAGUCAACUACUGCCAAUAUAUAUUUG--AAGCCUGAGGCAUUGAAUUAUGCCACAGGCUCA ((((.((((((((.(...(-(((((.((((......))).).))))))-.).))))))))..))))..........((--(.(((((.(((((......))))).)))))))) ( -36.10) >sy_au.0 808671 112 + 2809422/0-113 GGCAAGGUUGACUAGAAUUGAAAAAAGCUUGUUACAAGCGCAUUUUCG-UUCAGUCAACUACUGCCAAUAUAACUUUGUAGAGCAUUGAACAUUGAUUUAUGUCUCAAGCUCA ((((.((((((((.((((.(((((..(((((...)))))...))))))-)))))))))))..))))..............((((.((((((((......)))).)))))))). ( -38.80) >sy_sa.0 622241 110 + 2516575/0-113 GGCAAGGAUGACUAGGAUUGAAAAUAGCUUGAUAUAAGCGCAUUUUCACUUCAGUCAUCUACUGCCUAAAUGAAAAAG---GGUCUGAGACAUCUAUUUUUGUCUCAGACUCU ((((.((((((((.(((.(((((((.(((((...)))))..))))))).)))))))))))..))))..........((---(((((((((((........))))))))))))) ( -49.20) >consensus GGCAAGGUUGACUAGAAUUGAAAAAAGCUUGUUACAAGCGCAUUUUCA_UUCAGUCAACUACUGCCAAUAUAAAUUUG___AGCCUGAGACAUUGAUUUAUGUCUCAGGCUCA ((((.((((((((.(((.((((((..(((((...)))))...)))))).)))))))))))..))))...............((((((((((((......)))))))))))).. (-36.20 = -36.77 + 0.57)

| Location | 1,588,593 – 1,588,702 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 75.30 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -29.10 |
| Energy contribution | -31.00 |
| Covariance contribution | 1.90 |
| Combinations/Pair | 1.16 |
| Mean z-score | -5.18 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1588593 109 - 2685015/0-113 UGAGCCUGUGGCAUAAUUCAAUGCCUCAGGCUU--CAAAUAUAUAUUGGCAGUAGUUGACUGGC-UAAAAAUGCACUUGUAACAAUCUUUUUU-AAUUCUAGUCAACCUUGCC .(((((((.(((((......))))).)))))))--............(((((..(((((((((.-(((((((((....))).......)))))-)...))))))))).))))) ( -36.11) >sy_au.0 808671 112 + 2809422/0-113 UGAGCUUGAGACAUAAAUCAAUGUUCAAUGCUCUACAAAGUUAUAUUGGCAGUAGUUGACUGAA-CGAAAAUGCGCUUGUAACAAGCUUUUUUCAAUUCUAGUCAACCUUGCC .(((((((((.(((......)))))))).))))..............(((((..((((((((((-.(((((...(((((...)))))..)))))..))).))))))).))))) ( -32.50) >sy_sa.0 622241 110 + 2516575/0-113 AGAGUCUGAGACAAAAAUAGAUGUCUCAGACC---CUUUUUCAUUUAGGCAGUAGAUGACUGAAGUGAAAAUGCGCUUAUAUCAAGCUAUUUUCAAUCCUAGUCAUCCUUGCC ((.((((((((((........)))))))))).---))..........(((((..((((((((...((((((((.((((.....))))))))))))....)))))))).))))) ( -41.40) >consensus UGAGCCUGAGACAUAAAUCAAUGUCUCAGGCU___CAAAUUUAUAUUGGCAGUAGUUGACUGAA_UGAAAAUGCGCUUGUAACAAGCUUUUUUCAAUUCUAGUCAACCUUGCC .(((((((.(((((......))))).)))))))..............(((((..((((((((((.((((((...((((.....))))..)))))).))).))))))).))))) (-29.10 = -31.00 + 1.90) # Strand winner: forward (1.00)

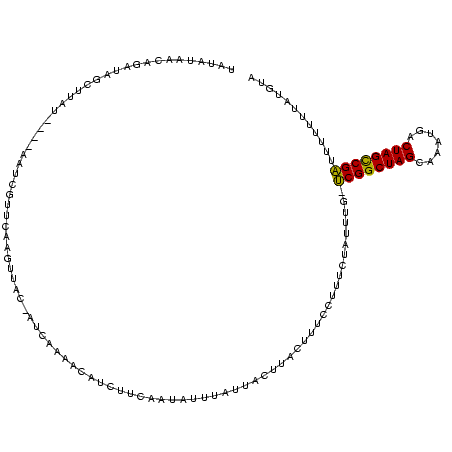
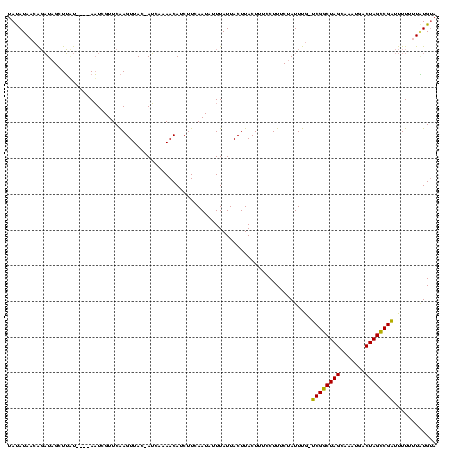
| Location | 765,300 – 765,418 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -19.42 |
| Consensus MFE | -13.13 |
| Energy contribution | -12.47 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
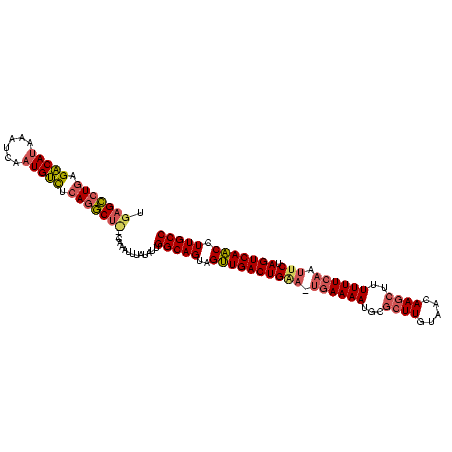
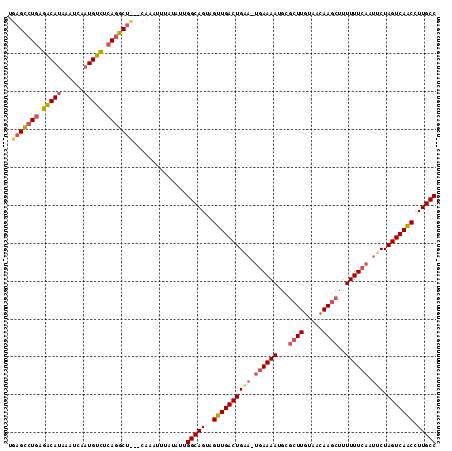
>sy_ha.0 765300 118 + 2685015/196-316 UAUAUAACAGAUAGCUUGUUGUGAAUCGUUCAAGUUAC-AUCGAAACAUCUUCAAUAUUUAUUACUUACUUUCCUUUCUAUUUG-UCGGCUAGCAGAUGACUAGCCGAUUUUUUUGUGUC ......((((((((((((.((.....))..))))))).-............................................(-(((((((((....).)))))))))...)))))... ( -21.50) >sy_au.0 442485 114 - 2809422/196-316 UAUAUAACAGAUAGCUUUU----AAUCGUUCAAGUUAC-AUCACAACAUCUUCAAUAUUUAUUACUUACUUUCCUUUCUAUUUG-UCGGCUAGCACGUGACUAGCCGAUUUUUUUAUGCA ..((((((((((((.....----....((..((((...-........................))))))........)))))))-(((((((((....).)))))))).....))))).. ( -16.16) >sy_sa.0 635941 118 + 2516575/196-316 UACAUAUAGUUUUGUUAACAUCUAUUUGUUCGAGUUACUAAUCAAACAUCUUCAAUAUUUAUUACUUACUUUCCUUUCUAUUUAACCGGCUAGCGAC--GCUAGUCGGUUUUAUUUUGUA ((((..((((..(...((((......))))...)..))))...........................................(((((((((((...--)))))))))))......)))) ( -20.60) >consensus UAUAUAACAGAUAGCUUAU____AAUCGUUCAAGUUAC_AUCAAAACAUCUUCAAUAUUUAUUACUUACUUUCCUUUCUAUUUG_UCGGCUAGCAAAUGACUAGCCGAUUUUUUUAUGUA .....................................................................................((((((((.......))))))))............ (-13.13 = -12.47 + -0.66)

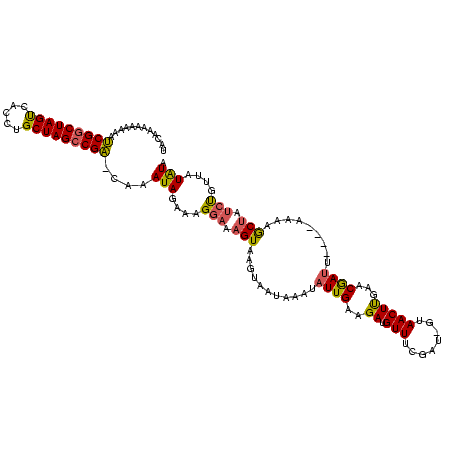
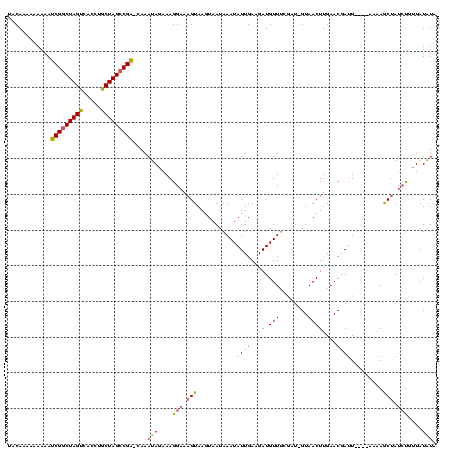
| Location | 765,300 – 765,418 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.26 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 765300 118 + 2685015/196-316 GACACAAAAAAAUCGGCUAGUCAUCUGCUAGCCGA-CAAAUAGAAAGGAAAGUAAGUAAUAAAUAUUGAAGAUGUUUCGAU-GUAACUUGAACGAUUCACAACAAGCUAUCUGUUAUAUA ............(((((((((.....)))))))))-..((((((.((...............(((((((((...)))))))-))..((((............)))))).))))))..... ( -24.60) >sy_au.0 442485 114 - 2809422/196-316 UGCAUAAAAAAAUCGGCUAGUCACGUGCUAGCCGA-CAAAUAGAAAGGAAAGUAAGUAAUAAAUAUUGAAGAUGUUGUGAU-GUAACUUGAACGAUU----AAAAGCUAUCUGUUAUAUA ............(((((((((.....)))))))))-..((((((.((.....(((((.((.((((((...))))))...))-...)))))..(....----....))).))))))..... ( -19.70) >sy_sa.0 635941 118 + 2516575/196-316 UACAAAAUAAAACCGACUAGC--GUCGCUAGCCGGUUAAAUAGAAAGGAAAGUAAGUAAUAAAUAUUGAAGAUGUUUGAUUAGUAACUCGAACAAAUAGAUGUUAACAAAACUAUAUGUA ((((......(((((.(((((--...))))).)))))........................((((((.....(((((((........)))))))....))))))............)))) ( -21.20) >consensus UACAAAAAAAAAUCGGCUAGUCACCUGCUAGCCGA_CAAAUAGAAAGGAAAGUAAGUAAUAAAUAUUGAAGAUGUUUCGAU_GUAACUUGAACGAUU____AAAAGCUAUCUGUUAUAUA ............(((((((((.....)))))))))....(((....(((.(((...........((((..((.(((........)))))...)))).........))).)))....))). (-13.88 = -13.78 + -0.10) # Strand winner: reverse (0.63)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:55 2006