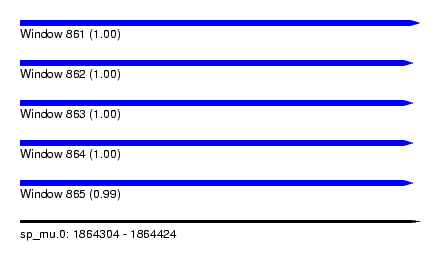
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 1,864,304 – 1,864,424 |
| Length | 120 |
| Max. P | 0.999932 |

| Location | 1,864,304 – 1,864,424 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -15.72 |
| Energy contribution | -13.85 |
| Covariance contribution | -1.87 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.57 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
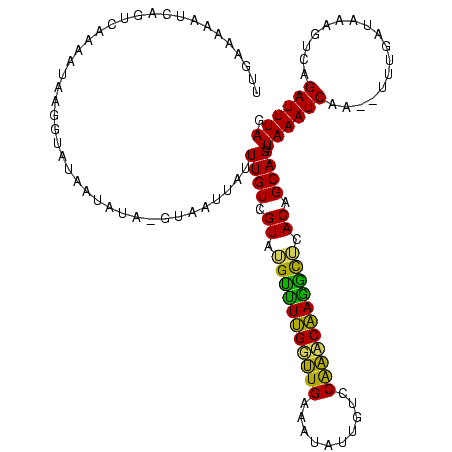
>sp_mu.0 1864304 120 + 2030921/0-120 UCCAAACAAAGCUCACAGCAGUUAAAUCAAGCUUUAAUGAAGUCCGAUUUAGCUGCUCUUUUUAUGCUUAAUUUUAGAAAAAAAGAUAAAUGUAAUCUAUAUUUAAAAAUUCUGAAAAUU ........((((....((((((((((((..(((((...)))))..))))))))))))........))))..((((((((.......((((((((...))))))))....))))))))... ( -26.60) >sp_ag.0 1981093 118 - 2160267/0-120 UCCGACCUAGAUAUACAGCAGUUAAAUCAA--CUUGAUAAAGUGAGAUUUAGCUGCUCUUUUUGUGCCUAAUUUUAAGAAAAAAGGUUAUUGUAAUAUAUAUUUAAACUUUCUGAAAAUU ...(((((.........(((((((((((.(--(((....))))..)))))))))))((((...((.....))...))))....)))))................................ ( -24.20) >sp_pn.0 1751729 117 + 2038615/0-120 UCCAGACAAGGCUCACAGCAGUUAAAUCU---UCUGAAAAAGUCAGAUUUAGCUGCUCUUUUUGUGCUUUUUUUCAAGAUUAUGAGCAUUUGUAACAGAGGCUUAAAGAUUCUGAAAAUU .......(((((.((.(((((((((((((---.((.....))..))))))))))))).....)).)))))((((((..(((.(((((.((((...)))).)))))..)))..)))))).. ( -32.00) >consensus UCCAAACAAGGCUCACAGCAGUUAAAUCAA__UUUGAUAAAGUCAGAUUUAGCUGCUCUUUUUGUGCUUAAUUUUAAGAAAAAAGGUAAUUGUAAUAUAUAUUUAAAAAUUCUGAAAAUU .......(((((....((((((((((((.................))))))))))))........))))).(((((.(((...((((.............))))....))).)))))... (-15.72 = -13.85 + -1.87) # Strand winner: reverse (0.99)

| Location | 1,864,304 – 1,864,422 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.76 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1864304 118 + 2030921/40-160 UUUCUAAAAUUAAGCAUAAAAAGAGCAGCUAAAUCGGACUUCAUUAAAGCUUGAUUUAACUGCUGUGAGCUUUGUUUGGACAAUAUUUCAACCAAAGCAAACGACAAAUAAUAA--UAAA .......................(((((.(((((((..(((.....)))..))))))).)))))((..((((((.(((((......))))).))))))..))............--.... ( -26.20) >sp_ag.0 1981093 118 - 2160267/40-160 UUCUUAAAAUUAGGCACAAAAAGAGCAGCUAAAUCUCACUUUAUCAAG--UUGAUUUAACUGCUGUAUAUCUAGGUCGGACAAUAUUUCAACCUAGACAGACGACAAAUAGUUACGUACA .......................(((((.((((((..((((....)))--).)))))).)))))((((.(((((((.(((......))).)))))))..(((........)))..)))). ( -26.30) >sp_pn.0 1751729 117 + 2038615/40-160 AUCUUGAAAAAAAGCACAAAAAGAGCAGCUAAAUCUGACUUUUUCAGA---AGAUUUAACUGCUGUGAGCCUUGUCUGGACAAUAUUUCAGACAAAACCUACGACAAAUGAUUACCCAUA ...(((.................(((((.(((((((..((.....)).---))))))).)))))(((.(..(((((((((......)))))))))..).)))..)))............. ( -24.60) >consensus UUCUUAAAAUUAAGCACAAAAAGAGCAGCUAAAUCUGACUUUAUCAAA__UUGAUUUAACUGCUGUGAGCCUUGUUUGGACAAUAUUUCAACCAAAACAAACGACAAAUAAUUAC_UAAA .......................(((((.((((((.................)))))).)))))((..((((((((((((......))))))))))))..)).................. (-22.64 = -22.76 + 0.12)

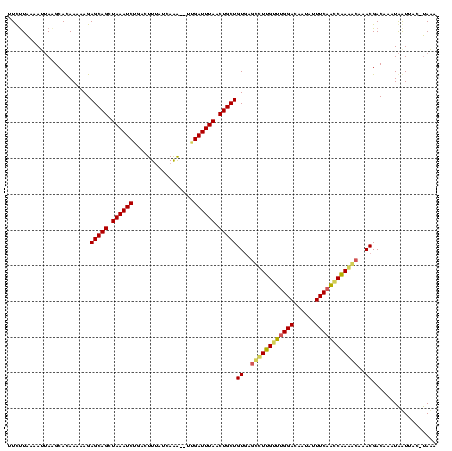
| Location | 1,864,304 – 1,864,422 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -29.73 |
| Energy contribution | -26.86 |
| Covariance contribution | -2.87 |
| Combinations/Pair | 1.47 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1864304 118 + 2030921/40-160 UUUA--UUAUUAUUUGUCGUUUGCUUUGGUUGAAAUAUUGUCCAAACAAAGCUCACAGCAGUUAAAUCAAGCUUUAAUGAAGUCCGAUUUAGCUGCUCUUUUUAUGCUUAAUUUUAGAAA ((((--..((((...((.((..((((((.(((..........))).))))))..))((((((((((((..(((((...)))))..))))))))))))........)).))))..)))).. ( -29.00) >sp_ag.0 1981093 118 - 2160267/40-160 UGUACGUAACUAUUUGUCGUCUGUCUAGGUUGAAAUAUUGUCCGACCUAGAUAUACAGCAGUUAAAUCAA--CUUGAUAAAGUGAGAUUUAGCUGCUCUUUUUGUGCCUAAUUUUAAGAA .(((((............((.(((((((((((..........))))))))))).))((((((((((((.(--(((....))))..)))))))))))).....)))))............. ( -38.10) >sp_pn.0 1751729 117 + 2038615/40-160 UAUGGGUAAUCAUUUGUCGUAGGUUUUGUCUGAAAUAUUGUCCAGACAAGGCUCACAGCAGUUAAAUCU---UCUGAAAAAGUCAGAUUUAGCUGCUCUUUUUGUGCUUUUUUUCAAGAU ...(((((..........((..((((((((((..........))))))))))..))(((((((((((((---.((.....))..))))))))))))).......)))))........... ( -33.90) >consensus UAUA_GUAAUUAUUUGUCGUAUGUUUUGGUUGAAAUAUUGUCCAAACAAGGCUCACAGCAGUUAAAUCAA__UUUGAUAAAGUCAGAUUUAGCUGCUCUUUUUGUGCUUAAUUUUAAGAA ..................((.(((((((((((..........))))))))))).))((((((((((((.................))))))))))))..(((((..........))))). (-29.73 = -26.86 + -2.87) # Strand winner: reverse (1.00)


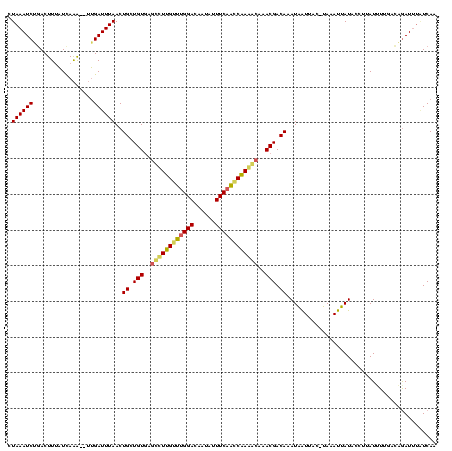
| Location | 1,864,304 – 1,864,422 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.35 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.06 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.72 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1864304 118 + 2030921/68-188 CUAAAUCGGACUUCAUUAAAGCUUGAUUUAACUGCUGUGAGCUUUGUUUGGACAAUAUUUCAACCAAAGCAAACGACAAAUAAUAA--UAAAUUAUACAUUAUAUUGACUGAUUUGUCAA .(((((((..(((.....)))..)))))))......((..((((((.(((((......))))).))))))..))(((((((..(((--((.(........).)))))....))))))).. ( -23.80) >sp_ag.0 1981093 118 - 2160267/68-188 CUAAAUCUCACUUUAUCAAG--UUGAUUUAACUGCUGUAUAUCUAGGUCGGACAAUAUUUCAACCUAGACAGACGACAAAUAGUUACGUACAUUAUAACUUAUUUUCACCAAUAAAUCAA ....................--((((((((......((...(((((((.(((......))).)))))))...))((.(((((((((.(.....).)))).))))))).....)))))))) ( -19.00) >sp_pn.0 1751729 117 + 2038615/68-188 CUAAAUCUGACUUUUUCAGA---AGAUUUAACUGCUGUGAGCCUUGUCUGGACAAUAUUUCAGACAAAACCUACGACAAAUGAUUACCCAUAUUAUACCCUAUUUAGCUAGAUUUUUCAA .....(((((.....)))))---(((((((.(((.((((.(..(((((((((......)))))))))..).))))....(((((.......)))))........))).)))))))..... ( -19.40) >consensus CUAAAUCUGACUUUAUCAAA__UUGAUUUAACUGCUGUGAGCCUUGUUUGGACAAUAUUUCAACCAAAACAAACGACAAAUAAUUAC_UAAAUUAUACCUUAUUUUGACAGAUUUAUCAA .((((((.................))))))..((.(((..((((((((((((......))))))))))))..))).)).......................................... (-14.94 = -15.06 + 0.12)

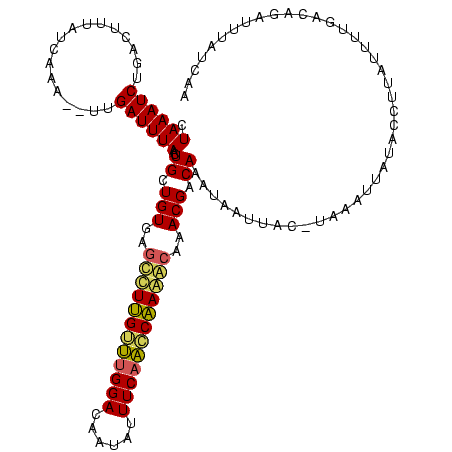
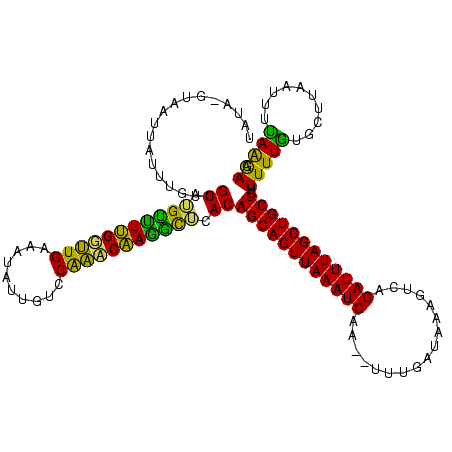
| Location | 1,864,304 – 1,864,422 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.35 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -19.62 |
| Energy contribution | -17.63 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1864304 118 + 2030921/68-188 UUGACAAAUCAGUCAAUAUAAUGUAUAAUUUA--UUAUUAUUUGUCGUUUGCUUUGGUUGAAAUAUUGUCCAAACAAAGCUCACAGCAGUUAAAUCAAGCUUUAAUGAAGUCCGAUUUAG (((((......))))).((((((.......))--))))...((((.((..((((((.(((..........))).))))))..)).)))).((((((..(((((...)))))..)))))). ( -21.70) >sp_ag.0 1981093 118 - 2160267/68-188 UUGAUUUAUUGGUGAAAAUAAGUUAUAAUGUACGUAACUAUUUGUCGUCUGUCUAGGUUGAAAUAUUGUCCGACCUAGAUAUACAGCAGUUAAAUCAA--CUUGAUAAAGUGAGAUUUAG .(((((((((......)))))))))................((((.((.(((((((((((..........))))))))))).)).)))).((((((.(--(((....))))..)))))). ( -29.50) >sp_pn.0 1751729 117 + 2038615/68-188 UUGAAAAAUCUAGCUAAAUAGGGUAUAAUAUGGGUAAUCAUUUGUCGUAGGUUUUGUCUGAAAUAUUGUCCAGACAAGGCUCACAGCAGUUAAAUCU---UCUGAAAAAGUCAGAUUUAG .....((((((..((....(((((.((((((((....)))).(((.((..((((((((((..........))))))))))..)).))))))).))))---).......))..)))))).. ( -26.00) >consensus UUGAAAAAUCAGUCAAAAUAAGGUAUAAUAUA_GUAAUUAUUUGUCGUAUGUUUUGGUUGAAAUAUUGUCCAAACAAGGCUCACAGCAGUUAAAUCAA__UUUGAUAAAGUCAGAUUUAG .........................................((((.((.(((((((((((..........))))))))))).)).)))).((((((.................)))))). (-19.62 = -17.63 + -1.99) # Strand winner: reverse (0.99)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:49:11 2006