
| Sequence ID | sp_pn.0 |
|---|---|
| Location | 1,704,345 – 1,704,428 |
| Length | 83 |
| Max. P | 0.999934 |

| Location | 1,704,345 – 1,704,428 |
|---|---|
| Length | 83 |
| Sequences | 3 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -15.21 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 4.66 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1704345 83 + 2038615 AUAGAUAUUAUUAGCACUCUUUGAACUGGAGUGCUAAUUCAUAAUUCUAUUGUAUCACUUGGUCAGAAAUAGUCAAGAAAAAA (((((....(((((((((((.......))))))))))).......))))).......(((((..(....)..)))))...... ( -22.40) >sp_mu.0 1835232 83 + 2030921 CCAUAGAUUUUUAGCACUCCAAUAUACUGAGUGCUAAUUACAUAUAUUAUUGUAUCACUUGGUCAAAAAUAGUCAAGAAAAAA .....((...(((((((((.........))))))))).((((........)))))).(((((..(....)..)))))...... ( -15.30) >sp_ag.0 2058164 82 + 2160267 UUUGAU-UUUUUAGCACUCUUAUACAGAGAGUGCUAACUACAAAAACUAUUCUAUCACUUGGUCAGAAUUAGUCAAGAGAAAA ((((((-(..(((((((((((.....)))))))))))...........((((((((....))).))))).)))))))...... ( -21.40) >consensus AUAGAUAUUUUUAGCACUCUUAUAAACAGAGUGCUAAUUACAAAUACUAUUGUAUCACUUGGUCAGAAAUAGUCAAGAAAAAA ..........((((((((((.......))))))))))....................(((((..(....)..)))))...... (-15.21 = -16.10 + 0.89)

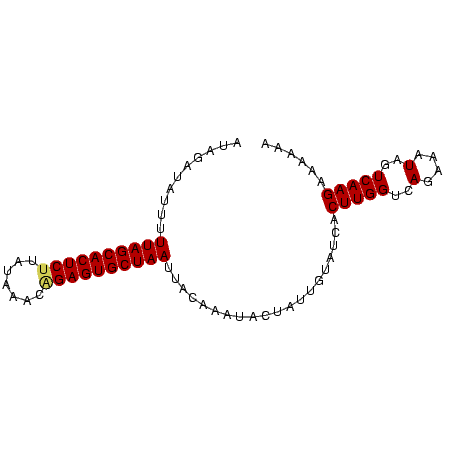
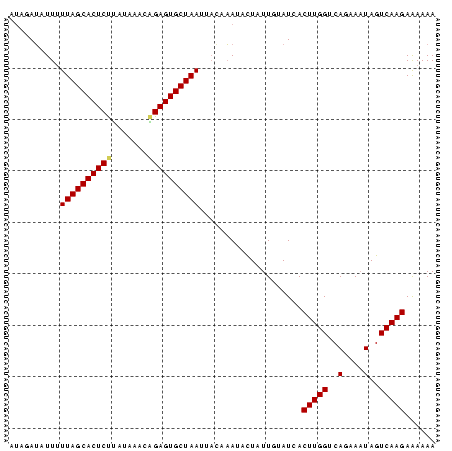
| Location | 1,704,345 – 1,704,428 |
|---|---|
| Length | 83 |
| Sequences | 3 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -16.53 |
| Consensus MFE | -13.42 |
| Energy contribution | -13.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1704345 83 + 2038615 UUUUUUCUUGACUAUUUCUGACCAAGUGAUACAAUAGAAUUAUGAAUUAGCACUCCAGUUCAAAGAGUGCUAAUAAUAUCUAU ....((((....((((.((.....)).))))....))))......((((((((((.........))))))))))......... ( -15.90) >sp_mu.0 1835232 83 + 2030921 UUUUUUCUUGACUAUUUUUGACCAAGUGAUACAAUAAUAUAUGUAAUUAGCACUCAGUAUAUUGGAGUGCUAAAAAUCUAUGG .....................(((.....((((........)))).(((((((((.........))))))))).......))) ( -14.90) >sp_ag.0 2058164 82 + 2160267 UUUUCUCUUGACUAAUUCUGACCAAGUGAUAGAAUAGUUUUUGUAGUUAGCACUCUCUGUAUAAGAGUGCUAAAAA-AUCAAA .........(((((.(((((.(.....).))))))))))((((...((((((((((.......))))))))))...-..)))) ( -18.80) >consensus UUUUUUCUUGACUAUUUCUGACCAAGUGAUACAAUAGUAUUUGUAAUUAGCACUCAAUAUAUAAGAGUGCUAAAAAUAUCUAA ...((.((((............)))).)).................(((((((((.........))))))))).......... (-13.42 = -13.20 + -0.22) # Strand winner: forward (0.91)

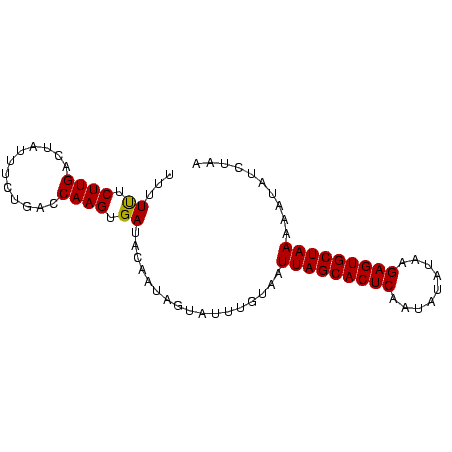
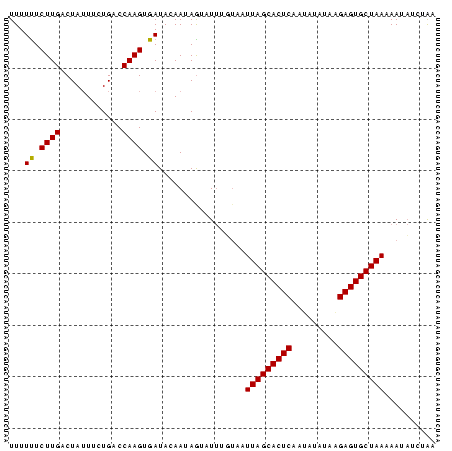
| Location | 1,704,345 – 1,704,428 |
|---|---|
| Length | 83 |
| Sequences | 3 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -15.21 |
| Energy contribution | -16.10 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 4.66 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1704345 83 + 2038615/0-83 AUAGAUAUUAUUAGCACUCUUUGAACUGGAGUGCUAAUUCAUAAUUCUAUUGUAUCACUUGGUCAGAAAUAGUCAAGAAAAAA (((((....(((((((((((.......))))))))))).......))))).......(((((..(....)..)))))...... ( -22.40) >sp_mu.0 1835232 83 + 2030921/0-83 CCAUAGAUUUUUAGCACUCCAAUAUACUGAGUGCUAAUUACAUAUAUUAUUGUAUCACUUGGUCAAAAAUAGUCAAGAAAAAA .....((...(((((((((.........))))))))).((((........)))))).(((((..(....)..)))))...... ( -15.30) >sp_ag.0 2058164 82 + 2160267/0-83 UUUGAU-UUUUUAGCACUCUUAUACAGAGAGUGCUAACUACAAAAACUAUUCUAUCACUUGGUCAGAAUUAGUCAAGAGAAAA ((((((-(..(((((((((((.....)))))))))))...........((((((((....))).))))).)))))))...... ( -21.40) >consensus AUAGAUAUUUUUAGCACUCUUAUAAACAGAGUGCUAAUUACAAAUACUAUUGUAUCACUUGGUCAGAAAUAGUCAAGAAAAAA ..........((((((((((.......))))))))))....................(((((..(....)..)))))...... (-15.21 = -16.10 + 0.89)

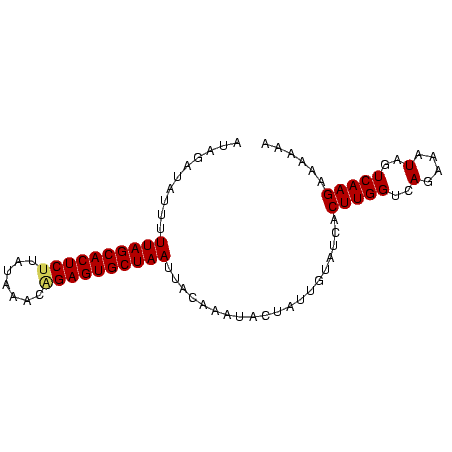
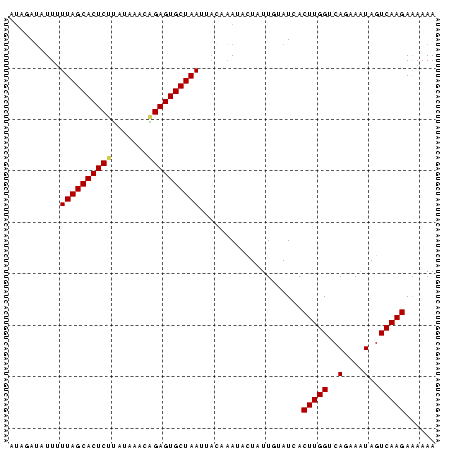
| Location | 1,704,345 – 1,704,428 |
|---|---|
| Length | 83 |
| Sequences | 3 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -16.53 |
| Consensus MFE | -13.42 |
| Energy contribution | -13.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1704345 83 + 2038615/0-83 UUUUUUCUUGACUAUUUCUGACCAAGUGAUACAAUAGAAUUAUGAAUUAGCACUCCAGUUCAAAGAGUGCUAAUAAUAUCUAU ....((((....((((.((.....)).))))....))))......((((((((((.........))))))))))......... ( -15.90) >sp_mu.0 1835232 83 + 2030921/0-83 UUUUUUCUUGACUAUUUUUGACCAAGUGAUACAAUAAUAUAUGUAAUUAGCACUCAGUAUAUUGGAGUGCUAAAAAUCUAUGG .....................(((.....((((........)))).(((((((((.........))))))))).......))) ( -14.90) >sp_ag.0 2058164 82 + 2160267/0-83 UUUUCUCUUGACUAAUUCUGACCAAGUGAUAGAAUAGUUUUUGUAGUUAGCACUCUCUGUAUAAGAGUGCUAAAAA-AUCAAA .........(((((.(((((.(.....).))))))))))((((...((((((((((.......))))))))))...-..)))) ( -18.80) >consensus UUUUUUCUUGACUAUUUCUGACCAAGUGAUACAAUAGUAUUUGUAAUUAGCACUCAAUAUAUAAGAGUGCUAAAAAUAUCUAA ...((.((((............)))).)).................(((((((((.........))))))))).......... (-13.42 = -13.20 + -0.22) # Strand winner: forward (0.91)

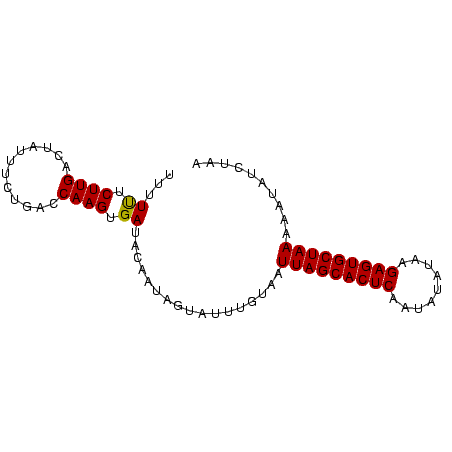
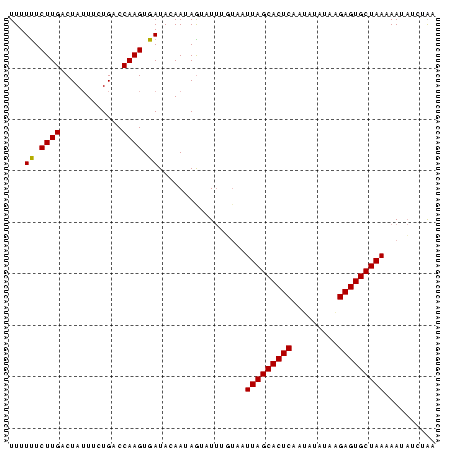
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:32 2006