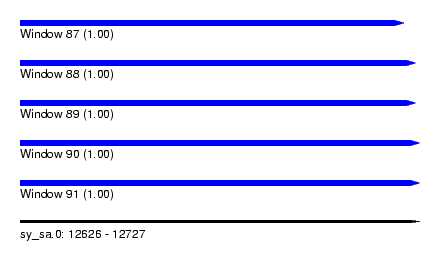
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 12,626 – 12,727 |
| Length | 101 |
| Max. P | 1.000000 |

| Location | 12,626 – 12,723 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.37 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.84 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 12626 97 + 2516575/120-240 AUAUUUAAUAUGAAUCUACCUACUAUAU----GAGAACAAUCGGUUAUAGCCGUUAUUU-AGUGAGAGUGCAGUCUAAUUUUGG------------------ACUGUUAAAUAGGGUGGC ...............((((((((((.((----((.......((((....)))))))).)-)))......((((((((....)))------------------)))))......)))))). ( -22.01) >sy_au.0 12463 119 + 2809422/120-240 AACAUAUAUAUGAAUCUACCUACUUAAUUUAAAAGAACAAUCGGUGAUAACCGUUAUUU-UAGUGAAGUGCAAUUUAGGUUUAGUGUAUCUUUAUAACUUAAAUUGUUAAAUAGGGUGGC ...............((((((((((...((((((.......((((....))))...)))-)))..))))((((((((((((.((.....))....))))))))))))......)))))). ( -25.70) >sy_ha.0 10997 108 + 2685015/120-240 GAUUGUUAUGCGAAUCUACCUACAAUU-----GUGAACAACCGGUAAUAACCGUUAUUUUAAGGAAAGUGCAAUUUGAUACAUAUGUA-------GUGUUAAAUUGUUAAAUAGGGUGGC ...............((((((((..((-----(....))).((((....))))..............))((((((((((((.......-------))))))))))))......)))))). ( -22.50) >consensus AAAUUUUAUAUGAAUCUACCUACUAUAU____GAGAACAAUCGGUAAUAACCGUUAUUU_AAGGAAAGUGCAAUUUAAUUUUAGUGUA__________UUAAAUUGUUAAAUAGGGUGGC ...............((((((....................((((....))))................((((((((((((..............))))))))))))......)))))). (-17.05 = -17.84 + 0.79)

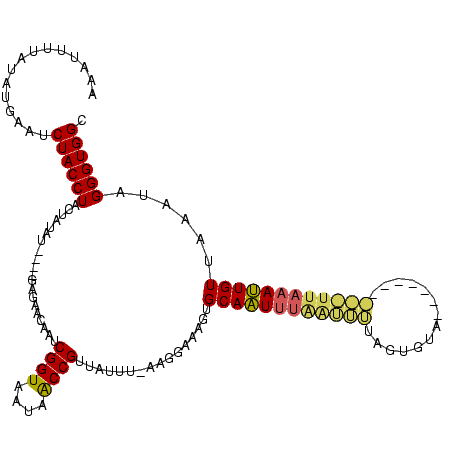
| Location | 12,626 – 12,726 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -37.17 |
| Energy contribution | -37.74 |
| Covariance contribution | 0.57 |
| Combinations/Pair | 1.26 |
| Mean z-score | -5.24 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 12626 100 + 2516575/160-280 UCGGUUAUAGCCGUUAUUU-AGUGAGAGUGCAGUCUAAUUUUGG------------------ACUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUACA-GGGACGUGGUCUU .((((....))))((((((-(........((((((((....)))------------------)))))))))))).(((....)))..(((((((((((((.....)-)))))))))))). ( -43.10) >sy_au.0 12463 116 + 2809422/160-280 UCGGUGAUAACCGUUAUUU-UAGUGAAGUGCAAUUUAGGUUUAGUGUAUCUUUAUAACUUAAAUUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUA---GGGAUGUGGUCUU ..(((....)))(((((((-((.......((((((((((((.((.....))....))))))))))))....))))))))).......((((((((((((.....---)))))))))))). ( -42.10) >sy_ha.0 10997 113 + 2685015/160-280 CCGGUAAUAACCGUUAUUUUAAGGAAAGUGCAAUUUGAUACAUAUGUA-------GUGUUAAAUUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUGAAGGGGAUGUGGUCUU (((((....))).................((((((((((((.......-------))))))))))))......))(((....)))..((((((((((((((....)))))))))))))). ( -44.30) >consensus UCGGUAAUAACCGUUAUUU_AAGGAAAGUGCAAUUUAAUUUUAGUGUA__________UUAAAUUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUA_A_GGGAUGUGGUCUU .((((....))))................((((((((((((..............))))))))))))........(((....)))..((((((((((((........)))))))))))). (-37.17 = -37.74 + 0.57)

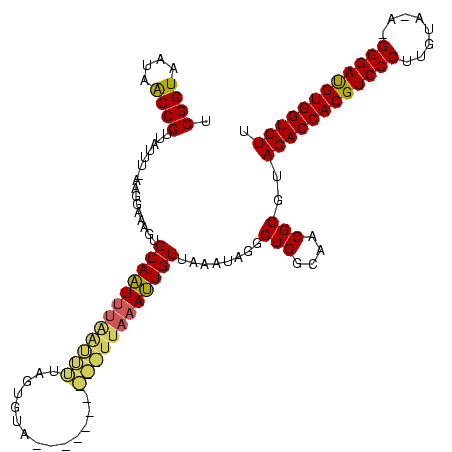
| Location | 12,626 – 12,726 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.22 |
| Mean single sequence MFE | -29.15 |
| Consensus MFE | -21.03 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.14 |
| Structure conservation index | 0.72 |
| SVM decision value | 6.15 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 12626 100 + 2516575/160-280 AAGACCACGUCCC-UGUACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAGU------------------CCAAAAUUAGACUGCACUCUCACU-AAAUAACGGCUAUAACCGA .((((((((((((-(.....))))))))))))).((.((((((....((((((.((((------------------(........))))).........)-)))))..)))...))))). ( -35.70) >sy_au.0 12463 116 + 2809422/160-280 AAGACCACAUCCC---UACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAAUUUAAGUUAUAAAGAUACACUAAACCUAAAUUGCACUUCACUA-AAAUAACGGUUAUCACCGA .(((((((.((((---.....)))).))))))).....................(((((((.(((..............))).)))))))..........-......((((....)))). ( -26.54) >sy_ha.0 10997 113 + 2685015/160-280 AAGACCACAUCCCCUUCACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAAUUUAACAC-------UACAUAUGUAUCAAAUUGCACUUUCCUUAAAAUAACGGUUAUUACCGG .(((((((.((((........)))).)))))))..((((((..........))))..........-------(((....))).......))................((((....)))). ( -25.20) >consensus AAGACCACAUCCC_U_UACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAAUUUAA__________UACACAAAAAUUAAAUUGCACUUUCCUU_AAAUAACGGUUAUAACCGA .(((((((.((((........)))).)))))))..........................................................................(((......))). (-21.03 = -21.03 + 0.00) # Strand winner: forward (1.00)

| Location | 12,626 – 12,727 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.05 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -32.95 |
| Energy contribution | -33.74 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.26 |
| Mean z-score | -6.10 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 12626 101 + 2516575/180-300 AGUGAGAGUGCAGUCUAAUUUUGG------------------ACUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUACA-GGGACGUGGUCUUUUUGUGUUAUUAUCCAAUAA .........((((((((....)))------------------)))))......((((((.((((((.(((((((((((((.....)-))))))))))))...)))))).))))))..... ( -47.00) >sy_au.0 12463 114 + 2809422/180-300 UAGUGAAGUGCAAUUUAGGUUUAGUGUAUCUUUAUAACUUAAAUUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUA---GGGAUGUGGUCUUUUU---UUAUUUUCUAAAAA .........((((((((((((.((.....))....))))))))))))....(((((((....)))..((((((((((((.....---))))))))))))....---......)))).... ( -38.30) >sy_ha.0 10997 107 + 2685015/180-300 AAGGAAAGUGCAAUUUGAUACAUAUGUA-------GUGUUAAAUUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUGAAGGGGAUGUGGUCUUUUU------UUAUUGAAAAU .........((((((((((((.......-------))))))))))))..(((((((((....)))..((((((((((((((....))))))))))))))...)------)))))...... ( -40.80) >consensus AAGGAAAGUGCAAUUUAAUUUUAGUGUA__________UUAAAUUGUUAAAUAGGGUGGCAACGCGUAGACCACGUCCCUUGUA_A_GGGAUGUGGUCUUUUU___UUAUUAUCCAAAAA .........((((((((((((..............))))))))))))........(((....)))..((((((((((((........))))))))))))..................... (-32.95 = -33.74 + 0.79)

| Location | 12,626 – 12,727 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.05 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -17.95 |
| Energy contribution | -17.73 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.14 |
| Structure conservation index | 0.70 |
| SVM decision value | 7.37 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 12626 101 + 2516575/180-300 UUAUUGGAUAAUAACACAAAAAGACCACGUCCC-UGUACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAGU------------------CCAAAAUUAGACUGCACUCUCACU ....(((....((((.(....((((((((((((-(.....)))))))))))))..).)))))))..........((((------------------(........))))).......... ( -33.20) >sy_au.0 12463 114 + 2809422/180-300 UUUUUAGAAAAUAA---AAAAAGACCACAUCCC---UACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAAUUUAAGUUAUAAAGAUACACUAAACCUAAAUUGCACUUCACUA ......(((.....---....(((((((.((((---.....)))).))))))).....................(((((((.(((..............))).)))))))...))).... ( -23.54) >sy_ha.0 10997 107 + 2685015/180-300 AUUUUCAAUAA------AAAAAGACCACAUCCCCUUCACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAAUUUAACAC-------UACAUAUGUAUCAAAUUGCACUUUCCUU ...........------....(((((((.((((........)))).)))))))..((((((..........))))..........-------(((....))).......))......... ( -20.30) >consensus UUUUUAGAUAAUAA___AAAAAGACCACAUCCC_U_UACAAGGGACGUGGUCUACGCGUUGCCACCCUAUUUAACAAUUUAA__________UACACAAAAAUUAAAUUGCACUUUCCUU ...((((..............(((((((.((((........)))).)))))))....((((..........)))).....................)))).................... (-17.95 = -17.73 + -0.22) # Strand winner: forward (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:42 2006