
| Sequence ID | sp_ag.0 |
|---|---|
| Location | 1,848,213 – 1,848,332 |
| Length | 119 |
| Max. P | 0.992793 |

| Location | 1,848,213 – 1,848,331 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -32.45 |
| Consensus MFE | -33.66 |
| Energy contribution | -25.75 |
| Covariance contribution | -7.91 |
| Combinations/Pair | 1.97 |
| Mean z-score | -1.45 |
| Structure conservation index | 1.04 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
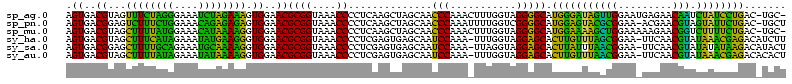
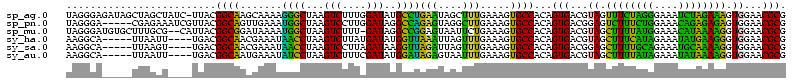
>sp_ag.0 1848213 118 - 2160267/120-240 AGUGACGUAGUUUCUAGGGAAAUCUAGAAAGUGGAACGCGGUAAACCCCUCAAGCUAGCAACCCAAACUUUGGUAGGGGCAUGGGAUAGUUGGAAUGAGAACAAUCUAUCCUGAC-UGC- .((..((...((((((((....)))))))).))..))(((((...(((((...(((((...........))))))))))...((((((((((.........))).))))))).))-)))- ( -32.80) >sp_pn.0 1701590 118 - 2038615/120-240 AGUGACGGAGUCUUUCUGGAAACAGAGAGAGUGGAACGCGGUAAACCCCUCAAGCUAGCAACCCAAAUUUUGGUCGGGGCAUGGAGUACGCGGAA-ACGAACGUAGUAUUCUGAC-UGCU .((..((...((((((((....)))))))).))..))(((((...((((....(....)...(((.....)))..))))...(((((((.((...-.))......))))))).))-))). ( -39.20) >sp_mu.0 1590727 118 - 2030921/120-240 AGUGACGUAGCUUUUAUGGAAACAUAAAAGGUGGAACGCGGUAAACCCCUCAAGCUAGCAACCCAAACUUUGGUAGGGGCAUGGAAAAGCUGGAAAAAGAACGGUCUUUUCUGAC-UGC- .((..((...((((((((....)))))))).))..))(((((...(((((...(((((...........))))))))))...((((((((((.........))).))))))).))-)))- ( -34.80) >sy_ha.0 1192558 118 - 2685015/120-240 AGUGACGUAGCUUUCAUAGAAAUAUGAAGGGUGGAACGCGGUAAACCCCUCGAGUGAGCAAUCCAAA-UUUGGUAGGAGCACUUGUUUAGCGGAA-UUCAACGUAUAAACGAGACAUCUU .(((...((.((((((((....)))))))).))...)))((....)).((((((((..(...(((..-..)))...)..)))))((((((((...-.....))).))))))))....... ( -32.00) >sy_sa.0 1173734 118 - 2516575/120-240 AGUGACGGAGCUUUUGCAGAAAUGCAAAAGGUGGAACGCGGUAAACCCCUCGAGUGAGCAAUCCAAA-UUAGGUAGGAGCACUUAUUUAACGGAA-UUCAACGUAUAUAUAAGACAUACU .(((....(.((((((((....)))))))).)....)))((((........(((((..(...((...-...))...)..))))).....(((...-.....)))............)))) ( -25.70) >sy_au.0 1525329 118 + 2809422/120-240 AGUGACGUAGCUUUUAUAGAAAUAUAAAAGGUGGAACGCGGUAAACCCCUCGAGUGAGCAAUCCAAA-UUUGGUAGGAGCACUUGUUUAACGGAA-UUCAACGUAUAAACGAGACACACU .(((...((.((((((((....)))))))).))......((....)).((((((((..(...(((..-..)))...)..)))))((((((((...-.....))).)))))))).)))... ( -30.20) >consensus AGUGACGUAGCUUUUAUAGAAAUAUAAAAGGUGGAACGCGGUAAACCCCUCAAGCGAGCAACCCAAA_UUUGGUAGGAGCACGGAUUAAACGGAA_UUCAACGUAUAAACCAGAC_UGCU .((..((...((((((((....)))))))).))..))((((....))..............(((...........))))).(((((((((((.........))).))))))))....... (-33.66 = -25.75 + -7.91) # Strand winner: reverse (1.00)

| Location | 1,848,213 – 1,848,332 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.27 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -16.99 |
| Energy contribution | -13.88 |
| Covariance contribution | -3.11 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 1848213 119 - 2160267/200-320 CGCGUUCCACUUUCUAGAUUUCCCUAGAAACUACGUCACUGUGGCACUUUCAAAGCUAUUCAGGCAUAUCCAAAGACUUAGCCCUUUUGCUUGCCGUAA-GAUAGCUAGCUAUCUCCCUA .((((.(((((((((((......)))))))..........)))).))......(((((((..((((...(.((((........)))).)..))))....-))))))).)).......... ( -27.20) >sp_pn.0 1701590 115 - 2038615/200-320 CGCGUUCCACUCUCUCUGUUUCCAGAAAGACUCCGUCACUGUGGCACUUUCAAGCCUACUCUGGCCUAUCCAAGGACUUAGCCAUUUCAACUGCCGUAACGAUUUCUCG-----UCCCUA .((..(((..(((.((((....)))).)))....((((..(((((........))).))..))))........)))....)).............(..((((....)))-----)..).. ( -22.00) >sp_mu.0 1590727 117 - 2030921/200-320 CGCGUUCCACCUUUUAUGUUUCCAUAAAAGCUACGUCACUGUGGCACUUUCAGAAUUACUCCGGCCUAUC-AAAGACUUAGCCAUUUUAUCCGCCGUAAUG--CGCAAAGCACAUCCCUA .((((...((.(((((((....)))))))((((.(((..((.(((..................)))...)-)..))).)))).............))...)--))).............. ( -20.47) >sy_ha.0 1192558 111 - 2685015/200-320 CGCGUUCCACCCUUCAUAUUUCUAUGAAAGCUACGUCACUGUGGCACUUUCAAACUAAUUUAACCAUAUCAUAAGACUUAGGUUAUUUCGUUGCCGUCA----AAUUAA-----UGCCUU .(((((......((((((....)))))).((((((....)))))).......(((.((..((((((..((....))..).))))).)).))).......----....))-----)))... ( -16.20) >sy_sa.0 1173734 111 - 2516575/200-320 CGCGUUCCACCUUUUGCAUUUCUGCAAAAGCUCCGUCACUGUGGCACUUUCAAACUAAUCUAACCUUAUCUAAGGACUUAGGUUAUUUCGUUGCCGUCA----ACUUAA-----UGCCUU .(((((....((((((((....)))))))).........(((((((....(.((.((((((((((((....))))..)))))))).)).).))))).))----....))-----)))... ( -24.70) >sy_au.0 1525329 111 + 2809422/200-320 CGCGUUCCACCUUUUAUAUUUCUAUAAAAGCUACGUCACUGUGGCACUUUCAAAUUACUCUAUCCAUAUCGAAAGACUUAGGAUAUUUCAUUGCCGUCA----AAUUAA-----UGCCUU .(((((.....(((((((....)))))))((((((....))))))...............((((((..((....))..).)))))..............----....))-----)))... ( -18.40) >consensus CGCGUUCCACCUUUUAUAUUUCCAUAAAAGCUACGUCACUGUGGCACUUUCAAACUAAUUCAGCCAUAUCCAAAGACUUAGCCUAUUUCAUUGCCGUAA____AACUAA_____UCCCUA .((((.((((((((((((....))))))))..........)))).))...............(((...((....))....))).........)).......................... (-16.99 = -13.88 + -3.11)

| Location | 1,848,213 – 1,848,332 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.27 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -27.99 |
| Energy contribution | -23.00 |
| Covariance contribution | -4.99 |
| Combinations/Pair | 1.85 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 1848213 119 - 2160267/200-320 UAGGGAGAUAGCUAGCUAUC-UUACGGCAAGCAAAAGGGCUAAGUCUUUGGAUAUGCCUGAAUAGCUUUGAAAGUGCCACAGUGACGUAGUUUCUAGGGAAAUCUAGAAAGUGGAACGCG ..(..((((((....)))))-)..)((((......(((((((..((...((.....)).)).))))))).....))))...(((...((.((((((((....)))))))).))...))). ( -35.10) >sp_pn.0 1701590 115 - 2038615/200-320 UAGGGA-----CGAGAAAUCGUUACGGCAGUUGAAAUGGCUAAGUCCUUGGAUAGGCCAGAGUAGGCUUGAAAGUGCCACAGUGACGGAGUCUUUCUGGAAACAGAGAGAGUGGAACGCG ..(..(-----(((....))))..)((((.((....(((((..(((....))).)))))(((....)))..)).))))...(((....(.((((((((....)))))))).)....))). ( -40.50) >sp_mu.0 1590727 117 - 2030921/200-320 UAGGGAUGUGCUUUGCG--CAUUACGGCGGAUAAAAUGGCUAAGUCUUU-GAUAGGCCGGAGUAAUUCUGAAAGUGCCACAGUGACGUAGCUUUUAUGGAAACAUAAAAGGUGGAACGCG ..(..((((((...)))--)))..)((((........((((..(((...-))).))))(((....)))......))))...(((...((.((((((((....)))))))).))...))). ( -33.80) >sy_ha.0 1192558 111 - 2685015/200-320 AAGGCA-----UUAAUU----UGACGGCAACGAAAUAACCUAAGUCUUAUGAUAUGGUUAAAUUAGUUUGAAAGUGCCACAGUGACGUAGCUUUCAUAGAAAUAUGAAGGGUGGAACGCG ..((((-----((....----...((....))...(((((...(((....)))..)))))............))))))...(((...((.((((((((....)))))))).))...))). ( -31.20) >sy_sa.0 1173734 111 - 2516575/200-320 AAGGCA-----UUAAGU----UGACGGCAACGAAAUAACCUAAGUCCUUAGAUAAGGUUAGAUUAGUUUGAAAGUGCCACAGUGACGGAGCUUUUGCAGAAAUGCAAAAGGUGGAACGCG ..((((-----((....----...((....))...((((((..(((....))).))))))............))))))...(((....(.((((((((....)))))))).)....))). ( -31.30) >sy_au.0 1525329 111 + 2809422/200-320 AAGGCA-----UUAAUU----UGACGGCAAUGAAAUAUCCUAAGUCUUUCGAUAUGGAUAGAGUAAUUUGAAAGUGCCACAGUGACGUAGCUUUUAUAGAAAUAUAAAAGGUGGAACGCG ..((((-----((...(----((....))).....(((((...(((....)))..)))))............))))))...(((...((.((((((((....)))))))).))...))). ( -24.70) >consensus AAGGCA_____UUAACU____UGACGGCAACGAAAAAGCCUAAGUCUUUGGAUAUGCCUAAAUAAGUUUGAAAGUGCCACAGUGACGUAGCUUUUAUAGAAAUAUAAAAGGUGGAACGCG .........................((((.......((((...(((....)))..))))(((....))).....))))...(((...((.((((((((....)))))))).))...))). (-27.99 = -23.00 + -4.99) # Strand winner: reverse (1.00)

| Location | 1,848,213 – 1,848,332 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.78 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -20.30 |
| Energy contribution | -16.47 |
| Covariance contribution | -3.83 |
| Combinations/Pair | 1.75 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
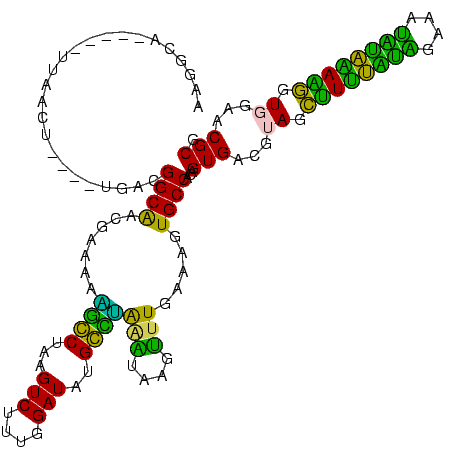
>sp_ag.0 1848213 119 - 2160267/240-360 GUGAUGCCAGUAGUGUUUGUGCUAGGCGAAAAAAUAAGCCUAGGGAGAUAGCUAGCUAUC-UUACGGCAAGCAAAAGGGCUAAGUCUUUGGAUAUGCCUGAAUAGCUUUGAAAGUGCCAC (((.((((.............((((((..........)))))).(((((((....)))))-))..)))).(((..(((((((..((...((.....)).)).))))))).....)))))) ( -35.60) >sp_pn.0 1701590 115 - 2038615/240-360 GUGAUGCCUGUAGUGUUUGUGCUAGGCGAAACCAUAAGCCUAGGGA-----CGAGAAAUCGUUACGGCAGUUGAAAUGGCUAAGUCCUUGGAUAGGCCAGAGUAGGCUUGAAAGUGCCAC ....((((.............((((((..........)))))).((-----(((....)))))..)))).......(((((..(((....))).))))).....(((........))).. ( -35.80) >sp_mu.0 1590727 117 - 2030921/240-360 GUGAUGCCAGUAGUGUUUGUGCUAGACAAAAAAAUAAGUCUAGGGAUGUGCUUUGCG--CAUUACGGCGGAUAAAAUGGCUAAGUCUUU-GAUAGGCCGGAGUAAUUCUGAAAGUGCCAC ((((((((((.((..(...(.((((((..........)))))).)..)..))))).)--))))))((((........((((..(((...-))).))))(((....)))......)))).. ( -34.30) >sy_ha.0 1192558 111 - 2685015/240-360 GAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAAUCAAGGCA-----UUAAUU----UGACGGCAACGAAAUAACCUAAGUCUUAUGAUAUGGUUAAAUUAGUUUGAAAGUGCCAC ....(((((....(((((((.....))).))))...))))).((((-----((....----...((....))...(((((...(((....)))..)))))............)))))).. ( -25.00) >sy_sa.0 1173734 111 - 2516575/240-360 GAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAGUCAAGGCA-----UUAAGU----UGACGGCAACGAAAUAACCUAAGUCCUUAGAUAAGGUUAGAUUAGUUUGAAAGUGCCAC ....(((((....(((((((.....))).))))...))))).((((-----((....----...((....))...((((((..(((....))).))))))............)))))).. ( -24.50) >sy_au.0 1525329 111 + 2809422/240-360 GAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAAUCAAGGCA-----UUAAUU----UGACGGCAAUGAAAUAUCCUAAGUCUUUCGAUAUGGAUAGAGUAAUUUGAAAGUGCCAC ....(((((....(((((((.....))).))))...))))).((((-----((...(----((....))).....(((((...(((....)))..)))))............)))))).. ( -20.50) >consensus GAGAUGACUGUAGUGUUCGUGCUAGACGAAACAAUAAGUCAAGGCA_____UUAACU____UGACGGCAACGAAAAAGCCUAAGUCUUUGGAUAUGCCUAAAUAAGUUUGAAAGUGCCAC .....................((((((..........))))))......................((((.......((((...(((....)))..))))(((....))).....)))).. (-20.30 = -16.47 + -3.83) # Strand winner: reverse (1.00)


| Location | 1,848,213 – 1,848,329 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.05 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -17.27 |
| Energy contribution | -15.38 |
| Covariance contribution | -1.89 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
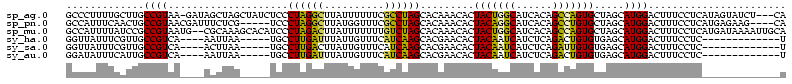
>sp_ag.0 1848213 116 - 2160267/280-400 GCCCUUUUGCUUGCCGUAA-GAUAGCUAGCUAUCUCCCUAGGCUUAUUUUUUCGCCUAGCACAAACACUACUGGCAUCACAGCCAGUGCUAGCAUGGACUUUCCUCAUAGUAUCU---CA ..((...((((.((.((.(-(((((....))))))..((((((..........)))))).)).......((((((......)))))))).)))).))(((........)))....---.. ( -32.10) >sp_pn.0 1701590 111 - 2038615/280-400 GCCAUUUCAACUGCCGUAACGAUUUCUCG-----UCCCUAGGCUUAUGGUUUCGCCUAGCACAAACACUACAGGCAUCACAGCCUGUGCUAGCAUGGACUUUCCUCAUGAGAAG----CA .......((...(((...((((....)))-----).....)))...))(((((..((((((((.........(((......)))))))))))(((((.......))))).))))----). ( -28.60) >sp_mu.0 1590727 118 - 2030921/280-400 GCCAUUUUAUCCGCCGUAAUG--CGCAAAGCACAUCCCUAGACUUAUUUUUUUGUCUAGCACAAACACUACUGGCAUCACAGCCAGUGCUAGCAUGGACUUUCCUCAUGAUAAAAUUGCA ((.((((((((.......(((--(....(((((....((((((..........))))))............((((......))))))))).))))((.....))....)))))))).)). ( -32.70) >sy_ha.0 1192558 98 - 2685015/280-400 GGUUAUUUCGUUGCCGUCA----AAUUAA-----UGCCUUGAUUUAUUGUUUCAUCAAGCACGAACACUACAAUCAUCUCAGACUGUGUGAGCAUGGACUUUCCUC-------------U (((.........)))(((.----.....(-----(((((((((..........))))))......(((.(((.((......)).)))))).)))).))).......-------------. ( -16.00) >sy_sa.0 1173734 98 - 2516575/280-400 GGUUAUUUCGUUGCCGUCA----ACUUAA-----UGCCUUGACUUAUUGUUUCAUCAAGCACGAACACUACAAUCAUCUCAGAUUGUGUGAGCAUGGACUUUCCUC-------------U .(((..(((((.((.((((----(.....-----....)))))...(((......))))))))))(((.((((((......))))))))))))..((.....))..-------------. ( -19.60) >sy_au.0 1525329 98 + 2809422/280-400 GGAUAUUUCAUUGCCGUCA----AAUUAA-----UGCCUUGAUUUAUUGUUUCAUCAAGCACGAACACUACAAUCAUCUCAGACUGUGUGAGCAUGGACUUUCCUC-------------U (((...(((((.((.....----......-----(((.(((((..........))))))))....(((.(((.((......)).)))))).)))))))...)))..-------------. ( -17.80) >consensus GCCUAUUUCAUUGCCGUAA____AACUAA_____UCCCUAGACUUAUUGUUUCAUCAAGCACAAACACUACAAGCAUCACAGACUGUGCGAGCAUGGACUUUCCUC_____________A .............((((....................((((((..........)))))).........(((((((......))))))).....))))....................... (-17.27 = -15.38 + -1.89)
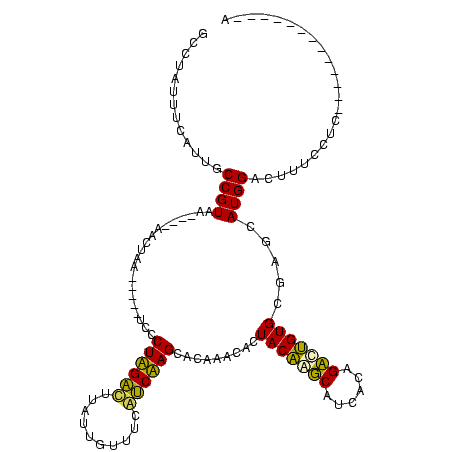
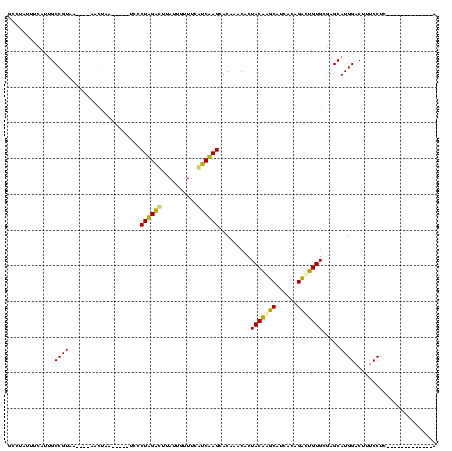
| Location | 1,848,213 – 1,848,329 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.05 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -23.85 |
| Energy contribution | -20.72 |
| Covariance contribution | -3.14 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989564 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 1848213 116 - 2160267/280-400 UG---AGAUACUAUGAGGAAAGUCCAUGCUAGCACUGGCUGUGAUGCCAGUAGUGUUUGUGCUAGGCGAAAAAAUAAGCCUAGGGAGAUAGCUAGCUAUC-UUACGGCAAGCAAAAGGGC ..---................((((........((((((......))))))..(((((((.((((((..........)))))).(((((((....)))))-))...)))))))...)))) ( -39.30) >sp_pn.0 1701590 111 - 2038615/280-400 UG----CUUCUCAUGAGGAAAGUCCAUGCUAGCACAGGCUGUGAUGCCUGUAGUGUUUGUGCUAGGCGAAACCAUAAGCCUAGGGA-----CGAGAAAUCGUUACGGCAGUUGAAAUGGC ..----((((....)))).....((((..((((((((((......))))))..((((.((.((((((..........)))))).((-----(((....)))))))))))))))..)))). ( -36.80) >sp_mu.0 1590727 118 - 2030921/280-400 UGCAAUUUUAUCAUGAGGAAAGUCCAUGCUAGCACUGGCUGUGAUGCCAGUAGUGUUUGUGCUAGACAAAAAAAUAAGUCUAGGGAUGUGCUUUGCG--CAUUACGGCGGAUAAAAUGGC .((.((((((((....((.....))............(((((((((((((.((..(...(.((((((..........)))))).)..)..))))).)--))))))))).)))))))).)) ( -38.90) >sy_ha.0 1192558 98 - 2685015/280-400 A-------------GAGGAAAGUCCAUGCUCACACAGUCUGAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAAUCAAGGCA-----UUAAUU----UGACGGCAACGAAAUAACC .-------------..((...(((((((..(((((((((......)))))).)))..))))((((((..........))))))...-----......----.)))(....).......)) ( -22.70) >sy_sa.0 1173734 98 - 2516575/280-400 A-------------GAGGAAAGUCCAUGCUCACACAAUCUGAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAGUCAAGGCA-----UUAAGU----UGACGGCAACGAAAUAACC .-------------..((...(((((((..(((((((((......)))))).)))..))))((((((..........))))))...-----......----.)))(....).......)) ( -22.40) >sy_au.0 1525329 98 + 2809422/280-400 A-------------GAGGAAAGUCCAUGCUCACACAGUCUGAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAAUCAAGGCA-----UUAAUU----UGACGGCAAUGAAAUAUCC .-------------.......(((((((..(((((((((......)))))).)))..))))((((((..........))))))...-----......----.)))............... ( -20.90) >consensus A_____________GAGGAAAGUCCAUGCUAACACAGGCUGAGAUGACUGUAGUGUUCGUGCUAGACGAAACAAUAAGUCAAGGCA_____UUAACU____UGACGGCAACGAAAAAGCC ................((.....)).((((.((((((((......)))))).)).......((((((..........))))))......................))))........... (-23.85 = -20.72 + -3.14) # Strand winner: reverse (1.00)
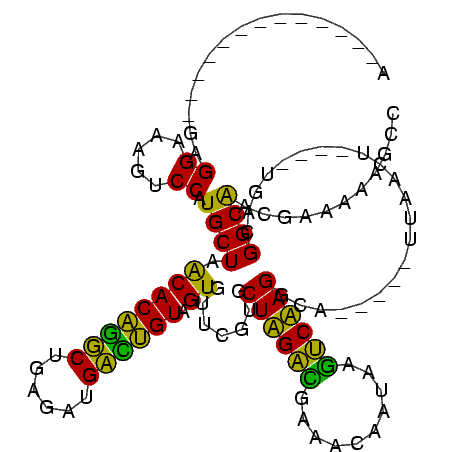

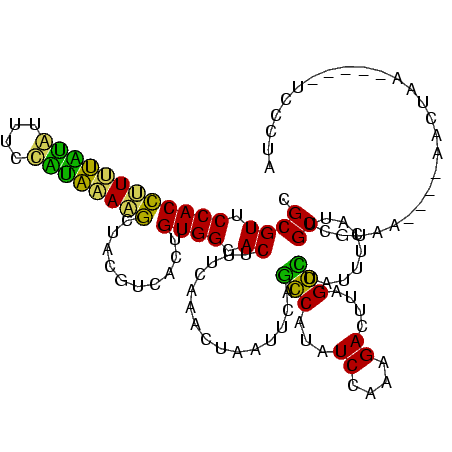
| Location | 1,848,213 – 1,848,326 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -15.52 |
| Energy contribution | -13.63 |
| Covariance contribution | -1.89 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.10 |
| SVM RNA-class probability | 0.988013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
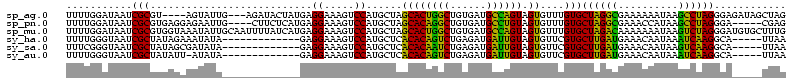
>sp_ag.0 1848213 113 - 2160267/305-425 CUAGCUAUCUCCCUAGGCUUAUUUUUUCGCCUAGCACAAACACUACUGGCAUCACAGCCAGUGCUAGCAUGGACUUUCCUCAUAGUAUCU---CAAUACU----ACGCGAUUAUCCAAAA (((((.......((((((..........))))))..........((((((......)))))))))))..((((...((..(.((((((..---..)))))----).).))...))))... ( -30.70) >sp_pn.0 1701590 111 - 2038615/305-425 CUCG-----UCCCUAGGCUUAUGGUUUCGCCUAGCACAAACACUACAGGCAUCACAGCCUGUGCUAGCAUGGACUUUCCUCAUGAGAAG----CAAUUCUCCUCACGCGAUUAUCCAAAA ...(-----((((((((((...((.....)).)))........(((((((......)))))))))))...)))).........(((((.----...)))))................... ( -29.60) >sp_mu.0 1590727 120 - 2030921/305-425 CAAAGCACAUCCCUAGACUUAUUUUUUUGUCUAGCACAAACACUACUGGCAUCACAGCCAGUGCUAGCAUGGACUUUCCUCAUGAUAAAAUUGCAAUAUUUACCACGCGAUUAUCCAAAA ...(((((....((((((..........))))))............((((......)))))))))..(((((.......)))))....((((((............))))))........ ( -25.30) >sy_ha.0 1192558 102 - 2685015/305-425 UUAA-----UGCCUUGAUUUAUUGUUUCAUCAAGCACGAACACUACAAUCAUCUCAGACUGUGUGAGCAUGGACUUUCCUC-------------UAUAUUUCUAUAGCGAUUACCCAAAA ...(-----(((((((((..........))))))......(((.(((.((......)).)))))).))))((....((..(-------------((((....))))).))....)).... ( -15.10) >sy_sa.0 1173734 102 - 2516575/305-425 UUAA-----UGCCUUGACUUAUUGUUUCAUCAAGCACGAACACUACAAUCAUCUCAGAUUGUGUGAGCAUGGACUUUCCUC-------------UAUAUCGCUAUAGCGAUUACCCGAAA ...(-----((((((((............)))))......(((.((((((......))))))))).))))((.....))..-------------...(((((....)))))......... ( -20.30) >sy_au.0 1525329 101 + 2809422/305-425 UUAA-----UGCCUUGAUUUAUUGUUUCAUCAAGCACGAACACUACAAUCAUCUCAGACUGUGUGAGCAUGGACUUUCCUC-------------UAUAU-AAUAUAGCGAUUACCCAAAA ...(-----(((((((((..........))))))......(((.(((.((......)).)))))).))))((....((..(-------------((((.-..))))).))....)).... ( -14.20) >consensus CUAA_____UCCCUAGACUUAUUGUUUCAUCAAGCACAAACACUACAAGCAUCACAGACUGUGCGAGCAUGGACUUUCCUC_____________AAUAUUUCUAAAGCGAUUACCCAAAA .............(((((..........)))))((........(((((((......))))))).......((.....))...........................))............ (-15.52 = -13.63 + -1.89)
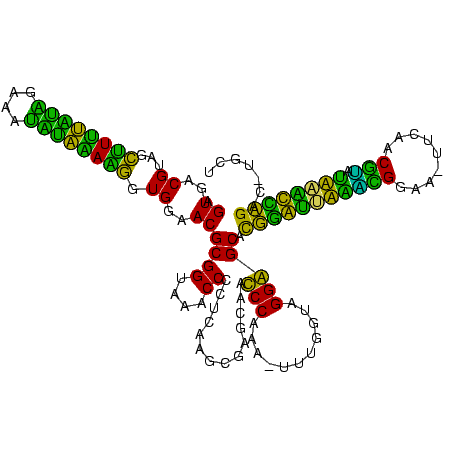
| Location | 1,848,213 – 1,848,326 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -21.43 |
| Energy contribution | -18.43 |
| Covariance contribution | -3.00 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
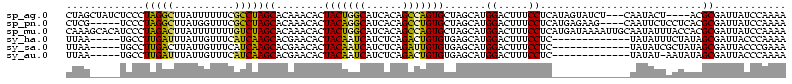
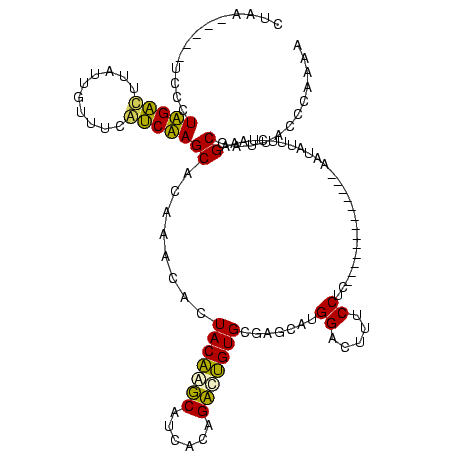
>sp_ag.0 1848213 113 - 2160267/305-425 UUUUGGAUAAUCGCGU----AGUAUUG---AGAUACUAUGAGGAAAGUCCAUGCUAGCACUGGCUGUGAUGCCAGUAGUGUUUGUGCUAGGCGAAAAAAUAAGCCUAGGGAGAUAGCUAG ...(((((..((.(((----(((((..---..))))))))..))..)))))..(((((((((((......))))))..(((((.(.((((((..........)))))).))))))))))) ( -41.80) >sp_pn.0 1701590 111 - 2038615/305-425 UUUUGGAUAAUCGCGUGAGGAGAAUUG----CUUCUCAUGAGGAAAGUCCAUGCUAGCACAGGCUGUGAUGCCUGUAGUGUUUGUGCUAGGCGAAACCAUAAGCCUAGGGA-----CGAG ...(((((..((.(((((((((.....----)))))))))..))..))))).....((((((((......)))))).))..((((.((((((..........)))))).).-----))). ( -40.10) >sp_mu.0 1590727 120 - 2030921/305-425 UUUUGGAUAAUCGCGUGGUAAAUAUUGCAAUUUUAUCAUGAGGAAAGUCCAUGCUAGCACUGGCUGUGAUGCCAGUAGUGUUUGUGCUAGACAAAAAAAUAAGUCUAGGGAUGUGCUUUG ...(((((..((.(((((((((.........)))))))))..))..))))).......((((((......))))))((..(...(.((((((..........)))))).)..)..))... ( -33.70) >sy_ha.0 1192558 102 - 2685015/305-425 UUUUGGGUAAUCGCUAUAGAAAUAUA-------------GAGGAAAGUCCAUGCUCACACAGUCUGAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAAUCAAGGCA-----UUAA ...((((...((.(((((....))))-------------)..))...))))(((.(((((((((......)))))).)))......((((((..........)))))))))-----.... ( -24.80) >sy_sa.0 1173734 102 - 2516575/305-425 UUUCGGGUAAUCGCUAUAGCGAUAUA-------------GAGGAAAGUCCAUGCUCACACAAUCUGAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAGUCAAGGCA-----UUAA ((((..(((.((((....))))))).-------------..))))....((((..(((((((((......)))))).)))..))))((((((..........))))))...-----.... ( -25.40) >sy_au.0 1525329 101 + 2809422/305-425 UUUUGGGUAAUCGCUAUAUU-AUAUA-------------GAGGAAAGUCCAUGCUCACACAGUCUGAGAUGAUUGUAGUGUUCGUGCUUGAUGAAACAAUAAAUCAAGGCA-----UUAA ...((((...((.(((((..-.))))-------------)..))...))))(((.(((((((((......)))))).)))......((((((..........)))))))))-----.... ( -21.10) >consensus UUUUGGAUAAUCGCGAUAGAAAUAUA_____________GAGGAAAGUCCAUGCUAACACAGGCUGAGAUGACUGUAGUGUUCGUGCUAGACGAAACAAUAAGUCAAGGCA_____UUAA ...........(((...........................((.....))......((((((((......)))))).))....)))((((((..........))))))............ (-21.43 = -18.43 + -3.00) # Strand winner: reverse (1.00)
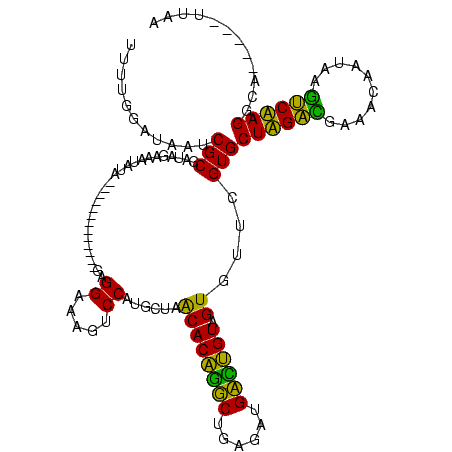
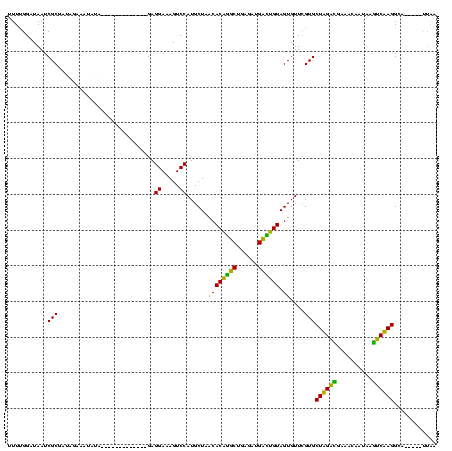
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:29 2006