
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 1,930,502 – 1,930,622 |
| Length | 120 |
| Max. P | 0.816172 |

| Location | 1,930,502 – 1,930,622 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -28.31 |
| Energy contribution | -27.87 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.816172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1930502 120 - 2030921/0-120 UUUGCGGAGAGAGGGACUUGAACCCUCACGACCAAAAGCGGUCACAAGAUCCUUAGUCUUGCGCGUCUGCCAAUUCCGCCAUCCCCGCGUAGCAACUUUUUAAAUUAUAUCAACUAACAA .((((((((((((((.......)))))..((((......)))).((((((.....))))))....))).)).....(((.......)))..))))......................... ( -28.10) >sp_ag.0 2041574 120 - 2160267/0-120 UCUGCGGAGAGAGGGACUUGAACCCUCACGACCAAAAGCGGUCACAAGAUCCUUAGUCUUGCGCGUCUGCCAAUUCCGCCAUCCCCGCUUAACAACAUAAUAAAUUAUAUCAACUUAUCA ...((((.(((((((.......)))))..((((......)))).((((((.....)))))).(((...........)))..)).))))................................ ( -27.50) >sp_pn.0 1421991 120 + 2038615/0-120 UAUGCGGAGAGAGGGACUUGAACCCUCACGACCUAAAGCGGUCACAGGAUCCUUAGUCCUGCACGUCUGCCAAUUCCGCCAUCCCCGCGUCGAUUACUUUACUAGUAUAUCAACUUUUGG .((((((.(((((((.......)))))..((((......)))).((((((.....))))))....................)).)))))).(((((((.....)))).)))......... ( -33.70) >consensus UAUGCGGAGAGAGGGACUUGAACCCUCACGACCAAAAGCGGUCACAAGAUCCUUAGUCUUGCGCGUCUGCCAAUUCCGCCAUCCCCGCGUAGCAACAUUUUAAAUUAUAUCAACUUAUAA ...((((((.(((((.......)))))..((((......)))).((((((.....))))))............))))))......................................... (-28.31 = -27.87 + -0.44)

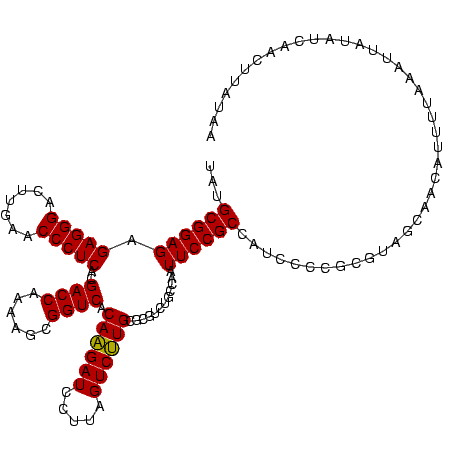
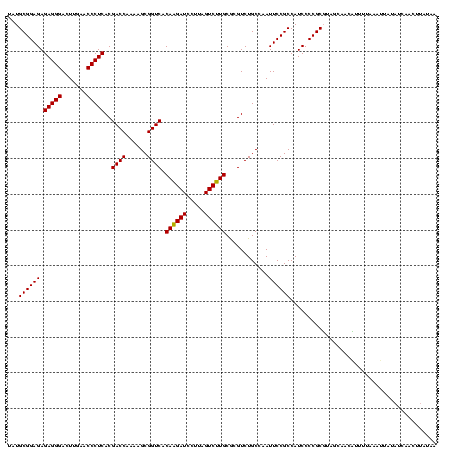
| Location | 1,930,502 – 1,930,622 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -35.43 |
| Energy contribution | -34.77 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804621 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1930502 120 - 2030921/0-120 UUGUUAGUUGAUAUAAUUUAAAAAGUUGCUACGCGGGGAUGGCGGAAUUGGCAGACGCGCAAGACUAAGGAUCUUGUGACCGCUUUUGGUCGUGAGGGUUCAAGUCCCUCUCUCCGCAAA ................................(((((((((((.(((..(((.(...((((((((....).))))))).).)))))).)))))(((((.......))))).))))))... ( -35.50) >sp_ag.0 2041574 120 - 2160267/0-120 UGAUAAGUUGAUAUAAUUUAUUAUGUUGUUAAGCGGGGAUGGCGGAAUUGGCAGACGCGCAAGACUAAGGAUCUUGUGACCGCUUUUGGUCGUGAGGGUUCAAGUCCCUCUCUCCGCAGA ((((((..(((((.....)))))..)))))).(((((((((((.(((..(((.(...((((((((....).))))))).).)))))).)))))(((((.......))))).))))))... ( -39.50) >sp_pn.0 1421991 120 + 2038615/0-120 CCAAAAGUUGAUAUACUAGUAAAGUAAUCGACGCGGGGAUGGCGGAAUUGGCAGACGUGCAGGACUAAGGAUCCUGUGACCGCUUUAGGUCGUGAGGGUUCAAGUCCCUCUCUCCGCAUA ......((((((.((((.....))))))))))(((((((.((........(((....))).(((((..(((((((((((((......)))))).))))))).))))))).)))))))... ( -44.20) >consensus UCAUAAGUUGAUAUAAUUUAAAAAGUUGCUACGCGGGGAUGGCGGAAUUGGCAGACGCGCAAGACUAAGGAUCUUGUGACCGCUUUUGGUCGUGAGGGUUCAAGUCCCUCUCUCCGCAAA ................................(((((((.(((((.....((....))(((((((....).))))))..))))).........(((((.......))))))))))))... (-35.43 = -34.77 + -0.66) # Strand winner: reverse (1.00)

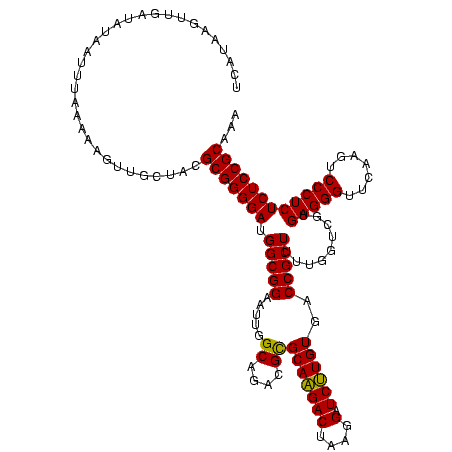
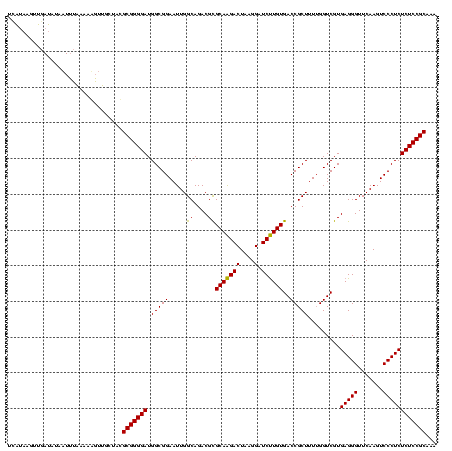
| Location | 1,930,502 – 1,930,622 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -25.61 |
| Energy contribution | -25.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1930502 120 - 2030921/12-132 UUGACACUAUCUUUGUUAGUUGAUAUAAUUUAAAAAGUUGCUACGCGGGGAUGGCGGAAUUGGCAGACGCGCAAGACUAAGGAUCUUGUGACCGCUUUUGGUCGUGAGGGUUCAAGUCCC ....(.((((((((((((((...................)))).)))))))))).)...(((((....)).)))((((..((((((..((((((....))))))..).))))).)))).. ( -30.81) >sp_ag.0 2041574 120 - 2160267/12-132 UUGACAACAAUAUGAUAAGUUGAUAUAAUUUAUUAUGUUGUUAAGCGGGGAUGGCGGAAUUGGCAGACGCGCAAGACUAAGGAUCUUGUGACCGCUUUUGGUCGUGAGGGUUCAAGUCCC .....(((((((((((((((((...)))))))))))))))))..(((.(..((.((....)).))..).)))..((((..((((((..((((((....))))))..).))))).)))).. ( -40.20) >sp_pn.0 1421991 120 + 2038615/12-132 UUGACAAGCAUCCCAAAAGUUGAUAUACUAGUAAAGUAAUCGACGCGGGGAUGGCGGAAUUGGCAGACGUGCAGGACUAAGGAUCCUGUGACCGCUUUAGGUCGUGAGGGUUCAAGUCCC ........((((((....((((((.((((.....))))))))))...)))))).........(((....))).(((((..(((((((((((((......)))))).))))))).))))). ( -41.90) >consensus UUGACAACAAUAUCAUAAGUUGAUAUAAUUUAAAAAGUUGCUACGCGGGGAUGGCGGAAUUGGCAGACGCGCAAGACUAAGGAUCUUGUGACCGCUUUUGGUCGUGAGGGUUCAAGUCCC ..(((.......................................(((.(..((.((....)).))..).)))........(((((((((((((......)))))).)))))))..))).. (-25.61 = -25.17 + -0.44) # Strand winner: reverse (1.00)

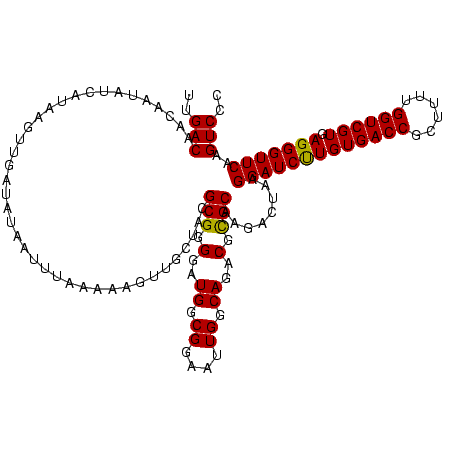
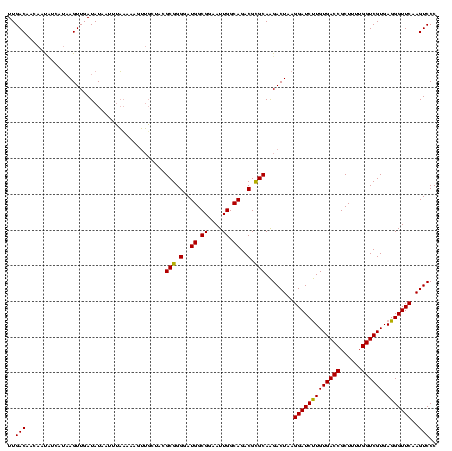
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:21 2006