| Sequence ID | sp_pn.0 |
|---|---|
| Location | 1,318,576 – 1,318,686 |
| Length | 110 |
| Max. P | 0.979466 |

| Location | 1,318,576 – 1,318,681 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 56.66 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -15.45 |
| Energy contribution | -11.70 |
| Covariance contribution | -3.75 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1318576 105 + 2038615/40-160 AGGAAAGAAAGUCUUAUGGC-GUUCUUCAAGCGAGCUUGGGAUAGUGGGAGCCAAGUAGGGCAAAAUAAAGGGCUGGCGCUUUCU--------GUAGUAUUUUCAAAAAC------AAUG .((((((((.(((....)))-.)))..((((....))))(((.((((..((((...((........))...))))..)))).)))--------......)))))......------.... ( -23.00) >sp_mu.0 1616400 119 - 2030921/40-160 AAGAAAGAGAGUUUUGUGGC-GUUUCUGCAGCGAACCUGAGAGAGUGUAAGUCAGGUGAAACAAAAUAAAGGACUGGCACUUUCUCUUGGCUAAUAGCCAAGCUAACAAUCAGAUAAAUG ..((.((((((((....)))-.)))))..(((......((((((((((.((((..((........))....)))).))))))))))(((((.....)))))))).....))......... ( -32.80) >sp_ag.0 1886578 110 - 2160267/40-160 AAGAAGAGGUGUUCAUCAGCUGUUUUUUUAGCGAGCUCAAGGUAGUGAAAGUUGGGUAGAAUAGGAUGAA-UAUUGGCACUUUUUAGUGAGUAAAAGUACAAUCAAA---------AAUG .....((((((((((((.((((......))))..((((((..(.....)..)))))).......))))))-))))..((((....)))).............))...---------.... ( -21.80) >consensus AAGAAAGAGAGUUUUAUGGC_GUUUUUCAAGCGAGCUUGAGAUAGUGAAAGUCAGGUAGAACAAAAUAAAGGACUGGCACUUUCU__UG__UAAUAGUAAAAUCAAAAA_______AAUG .((((((..((((((.((...(((((........((((((...........)))))))))))....)).))))))....))))))................................... (-15.45 = -11.70 + -3.75) # Strand winner: reverse (0.98)

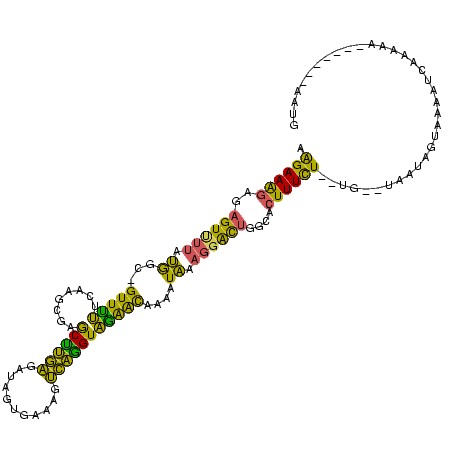
| Location | 1,318,576 – 1,318,686 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.89 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -15.45 |
| Energy contribution | -11.70 |
| Covariance contribution | -3.75 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1318576 110 + 2038615/61-181 UUUUUGAUAUAAUAG-UCAGCAGGAAAGAAAGUCUUAUGGC-GUUCUUCAAGCGAGCUUGGGAUAGUGGGAGCCAAGUAGGGCAAAAUAAAGGGCUGGCGCUUUCU--------GUAGUA ....((((......)-)))((..(((.(((.(((....)))-.))))))..))..(((..(((.((((..((((...((........))...))))..)))).)))--------..))). ( -26.50) >sp_mu.0 1616400 118 - 2030921/61-181 UUUUUGCUAUAAUCU-UUGUUAAGAAAGAGAGUUUUGUGGC-GUUUCUGCAGCGAACCUGAGAGAGUGUAAGUCAGGUGAAACAAAAUAAAGGACUGGCACUUUCUCUUGGCUAAUAGCC .....(((((..(((-((........)))))..(((((.((-(....))).)))))((.((((((((((.((((..((........))....)))).))))))))))..))...))))). ( -33.10) >sp_ag.0 1886578 119 - 2160267/61-181 UUUUUGUUAUACUGUCUCAGUAAGAAGAGGUGUUCAUCAGCUGUUUUUUUAGCGAGCUCAAGGUAGUGAAAGUUGGGUAGAAUAGGAUGAA-UAUUGGCACUUUUUAGUGAGUAAAAGUA .........((((..((((.(((((((.((((((((((.((((......))))..((((((..(.....)..)))))).......))))))-))))....))))))).))))....)))) ( -32.80) >consensus UUUUUGAUAUAAUAU_UCAGUAAGAAAGAGAGUUUUAUGGC_GUUUUUCAAGCGAGCUUGAGAUAGUGAAAGUCAGGUAGAACAAAAUAAAGGACUGGCACUUUCU__UG__UAAUAGUA ......................((((((..((((((.((...(((((........((((((...........)))))))))))....)).))))))....)))))).............. (-15.45 = -11.70 + -3.75) # Strand winner: reverse (1.00)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:19 2006