| Sequence ID | sp_pn.0 |
|---|---|
| Location | 1,242,028 – 1,242,146 |
| Length | 118 |
| Max. P | 0.999256 |

| Location | 1,242,028 – 1,242,146 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -20.13 |
| Energy contribution | -21.47 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995816 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1242028 118 + 2038615/0-120 CAAAAGACAAGCUCAUAUCACGAAGGGCGAA-AAACCGCGGUACCACCUUCAUUCAAUGAACUUGUCAUUCUCUUGUUCUUAUGCAAUUGUAUGAUUGAGUAGCAUGACUUCCUAGCU-U .....((((((.((((.....((((((((..-....)))(....).))))).....)))).))))))....((.((((((((..((......))..)))).)))).))..........-. ( -26.80) >sp_ag.0 1351795 116 - 2160267/0-120 CAAAAGACAAGUCCAU--UACGAAGGGCGAA-AA-CCGCGGUACCACCUUCAUUCAAUGAACUUGUCAUUCUCAUUUAUUUUUAGUUUUACCCCAUUGAGUAGCAUCUUUUUCUGAUUAU .....(((((((.(((--(..((((((((..-..-.)))(....).)))))....)))).)))))))...((((......................)))).................... ( -22.75) >sp_mu.0 1422527 119 - 2030921/0-120 CAAAAAACAAGUUCACUGUAAGAAGGGCGAAGAAACCGCGGUACCACCUUCAUUCAAUGAACUUGUCAUUCUUAUUAUGUUAUUCUGUUUAUCAAUCAAGCAGCCAAAUUUUUCUAUU-U ......(((((((((.((...((((((((.......)))(....).)))))...)).)))))))))..................((((((.......))))))...............-. ( -25.30) >consensus CAAAAGACAAGUUCAU__UACGAAGGGCGAA_AAACCGCGGUACCACCUUCAUUCAAUGAACUUGUCAUUCUCAUUUUCUUAUACUAUUAAACAAUUGAGUAGCAUAAUUUUCUAAUU_U .....(((((((((((.....((((((((.......)))(....).))))).....)))))))))))..................................................... (-20.13 = -21.47 + 1.33)

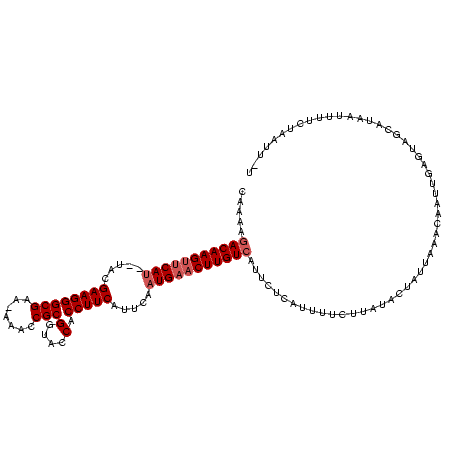
| Location | 1,242,028 – 1,242,146 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.02 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.20 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1242028 118 + 2038615/0-120 A-AGCUAGGAAGUCAUGCUACUCAAUCAUACAAUUGCAUAAGAACAAGAGAAUGACAAGUUCAUUGAAUGAAGGUGGUACCGCGGUUU-UUCGCCCUUCGUGAUAUGAGCUUGUCUUUUG .-............((((.................)))).........((((.(((((((((((...(((((((.(....)((((...-.)))))))))))...))))))))))))))). ( -31.23) >sp_ag.0 1351795 116 - 2160267/0-120 AUAAUCAGAAAAAGAUGCUACUCAAUGGGGUAAAACUAAAAAUAAAUGAGAAUGACAAGUUCAUUGAAUGAAGGUGGUACCGCGG-UU-UUCGCCCUUCGUA--AUGGACUUGUCUUUUG .....(((((........(((((....))))).....................((((((((((((..((((((((((.((....)-).-.))).))))))))--)))))))))))))))) ( -30.20) >sp_mu.0 1422527 119 - 2030921/0-120 A-AAUAGAAAAAUUUGGCUGCUUGAUUGAUAAACAGAAUAACAUAAUAAGAAUGACAAGUUCAUUGAAUGAAGGUGGUACCGCGGUUUCUUCGCCCUUCUUACAGUGAACUUGUUUUUUG .-..........((((..(((......).))..))))............(((.(((((((((((((...(((((.(....)((((.....)))))))))...))))))))))))).))). ( -28.80) >consensus A_AAUAAGAAAAUCAUGCUACUCAAUCGAAAAAAAGAAUAAAAAAAUGAGAAUGACAAGUUCAUUGAAUGAAGGUGGUACCGCGGUUU_UUCGCCCUUCGUA__AUGAACUUGUCUUUUG .................................................(((.(((((((((((...(((((((.(....)((((.....)))))))))))...))))))))))).))). (-26.75 = -26.20 + -0.55) # Strand winner: reverse (1.00)



Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:17 2006