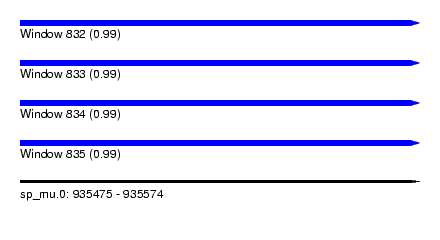
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 935,475 – 935,574 |
| Length | 99 |
| Max. P | 0.990460 |

| Location | 935,475 – 935,574 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -24.81 |
| Energy contribution | -23.43 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 935475 99 + 2030921 AAACUAUGCGAUGAGUCGAU-UGUGGUUUAU--ACCACAUACG--C-UAAGGAGAUCAUAA-UGCAGGAGCAGAUCUUGGCAAGUUGUGUGAACCUGCUUGCCACA .............((.((..-((((((....--))))))..))--)-)...((((((....-(((....)))))))))(((((((.(.(....)).)))))))... ( -29.20) >sp_ag.0 1096970 101 - 2160267 CUGUUAUGCGAUGAGUCGAUAUGUGGUUU-U--ACCACUAUUG--CGCAGGGAGAUUAUAAACGCAGGAGCGGAUCUUGAUAAGUUGUGUGAACCUUCUUGUCACA ..............(.(((((.((((...-.--.)))))))))--)((((((((.((((((((.(((((.....)))))....))).)))))..)))))))).... ( -24.40) >sp_pn.0 905032 105 + 2038615 AGAAUAUGCGAUGAGUCGAUAGGUAGUCUUCGGACUACUAUUGAGCAUAAGGAGGUCAUAA-CGCAGGAGCGGACCUUGAUGAGUUGUGUGAACCUGCUCAUCACA ....(((((......(((((((.(((((....)))))))))))))))))..((((((....-(((....)))))))))(((((((.(.(....)).)))))))... ( -39.30) >consensus AAAAUAUGCGAUGAGUCGAUAUGUGGUUU_U__ACCACUAUUG__C_UAAGGAGAUCAUAA_CGCAGGAGCGGAUCUUGAUAAGUUGUGUGAACCUGCUUGUCACA ........(((....)))....(((((......))))).............((((((.....(((....)))))))))((((((..(.(....))..))))))... (-24.81 = -23.43 + -1.38)

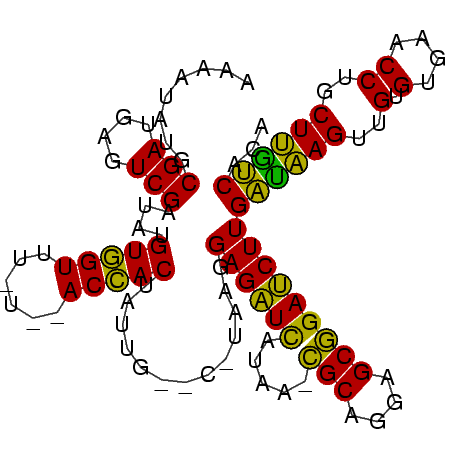
| Location | 935,475 – 935,574 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -20.27 |
| Energy contribution | -18.50 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 935475 99 + 2030921 UGUGGCAAGCAGGUUCACACAACUUGCCAAGAUCUGCUCCUGCA-UUAUGAUCUCCUUA-G--CGUAUGUGGU--AUAAACCACA-AUCGACUCAUCGCAUAGUUU ((((((..(((((((.....)))))))..((((((((....)))-....))))).....-)--)...((((((--....))))))-..........))))...... ( -25.20) >sp_ag.0 1096970 101 - 2160267 UGUGACAAGAAGGUUCACACAACUUAUCAAGAUCCGCUCCUGCGUUUAUAAUCUCCCUGCG--CAAUAGUGGU--A-AAACCACAUAUCGACUCAUCGCAUAACAG (((((..((.(((((.....)))))........(((((..(((((.............)))--))..))))).--.-..............))..)))))...... ( -18.42) >sp_pn.0 905032 105 + 2038615 UGUGAUGAGCAGGUUCACACAACUCAUCAAGGUCCGCUCCUGCG-UUAUGACCUCCUUAUGCUCAAUAGUAGUCCGAAGACUACCUAUCGACUCAUCGCAUAUUCU (((((((((..(((...............((((((((....)))-....)))))..............((((((....))))))..)))..)))))))))...... ( -29.80) >consensus UGUGACAAGCAGGUUCACACAACUUAUCAAGAUCCGCUCCUGCG_UUAUGAUCUCCUUA_G__CAAUAGUGGU__A_AAACCACAUAUCGACUCAUCGCAUAAUAU ..(((((((..(.......)..)))))))((((((((....))).....)))))..............(((((......)))))...................... (-20.27 = -18.50 + -1.77) # Strand winner: forward (1.00)

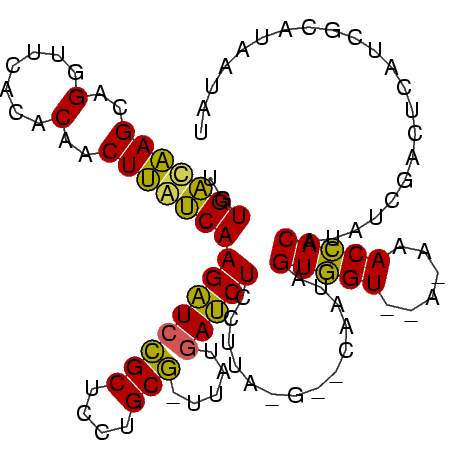
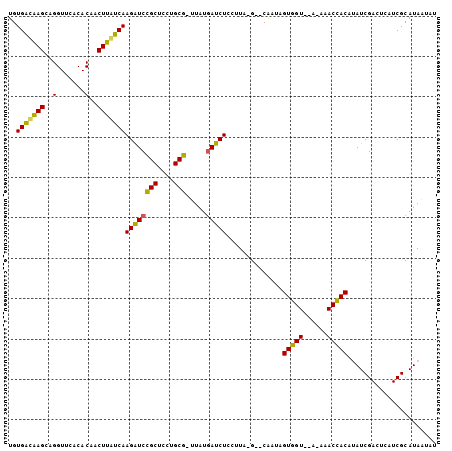
| Location | 935,475 – 935,574 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -24.81 |
| Energy contribution | -23.43 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 935475 99 + 2030921/0-106 AAACUAUGCGAUGAGUCGAU-UGUGGUUUAU--ACCACAUACG--C-UAAGGAGAUCAUAA-UGCAGGAGCAGAUCUUGGCAAGUUGUGUGAACCUGCUUGCCACA .............((.((..-((((((....--))))))..))--)-)...((((((....-(((....)))))))))(((((((.(.(....)).)))))))... ( -29.20) >sp_ag.0 1096970 101 - 2160267/0-106 CUGUUAUGCGAUGAGUCGAUAUGUGGUUU-U--ACCACUAUUG--CGCAGGGAGAUUAUAAACGCAGGAGCGGAUCUUGAUAAGUUGUGUGAACCUUCUUGUCACA ..............(.(((((.((((...-.--.)))))))))--)((((((((.((((((((.(((((.....)))))....))).)))))..)))))))).... ( -24.40) >sp_pn.0 905032 105 + 2038615/0-106 AGAAUAUGCGAUGAGUCGAUAGGUAGUCUUCGGACUACUAUUGAGCAUAAGGAGGUCAUAA-CGCAGGAGCGGACCUUGAUGAGUUGUGUGAACCUGCUCAUCACA ....(((((......(((((((.(((((....)))))))))))))))))..((((((....-(((....)))))))))(((((((.(.(....)).)))))))... ( -39.30) >consensus AAAAUAUGCGAUGAGUCGAUAUGUGGUUU_U__ACCACUAUUG__C_UAAGGAGAUCAUAA_CGCAGGAGCGGAUCUUGAUAAGUUGUGUGAACCUGCUUGUCACA ........(((....)))....(((((......))))).............((((((.....(((....)))))))))((((((..(.(....))..))))))... (-24.81 = -23.43 + -1.38)

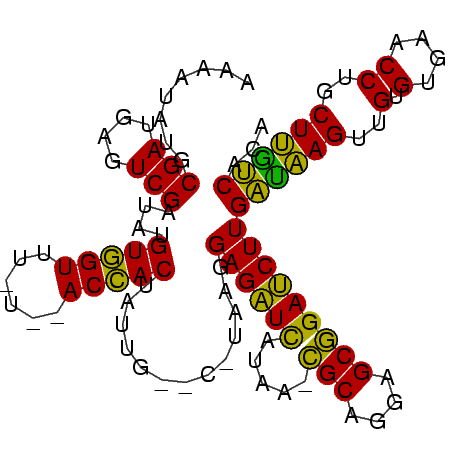
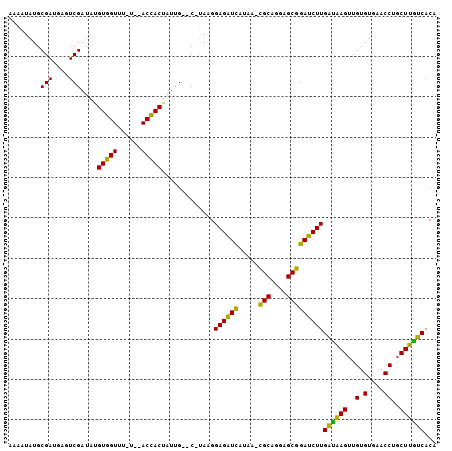
| Location | 935,475 – 935,574 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.36 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -20.27 |
| Energy contribution | -18.50 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 935475 99 + 2030921/0-106 UGUGGCAAGCAGGUUCACACAACUUGCCAAGAUCUGCUCCUGCA-UUAUGAUCUCCUUA-G--CGUAUGUGGU--AUAAACCACA-AUCGACUCAUCGCAUAGUUU ((((((..(((((((.....)))))))..((((((((....)))-....))))).....-)--)...((((((--....))))))-..........))))...... ( -25.20) >sp_ag.0 1096970 101 - 2160267/0-106 UGUGACAAGAAGGUUCACACAACUUAUCAAGAUCCGCUCCUGCGUUUAUAAUCUCCCUGCG--CAAUAGUGGU--A-AAACCACAUAUCGACUCAUCGCAUAACAG (((((..((.(((((.....)))))........(((((..(((((.............)))--))..))))).--.-..............))..)))))...... ( -18.42) >sp_pn.0 905032 105 + 2038615/0-106 UGUGAUGAGCAGGUUCACACAACUCAUCAAGGUCCGCUCCUGCG-UUAUGACCUCCUUAUGCUCAAUAGUAGUCCGAAGACUACCUAUCGACUCAUCGCAUAUUCU (((((((((..(((...............((((((((....)))-....)))))..............((((((....))))))..)))..)))))))))...... ( -29.80) >consensus UGUGACAAGCAGGUUCACACAACUUAUCAAGAUCCGCUCCUGCG_UUAUGAUCUCCUUA_G__CAAUAGUGGU__A_AAACCACAUAUCGACUCAUCGCAUAAUAU ..(((((((..(.......)..)))))))((((((((....))).....)))))..............(((((......)))))...................... (-20.27 = -18.50 + -1.77) # Strand winner: forward (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:13 2006