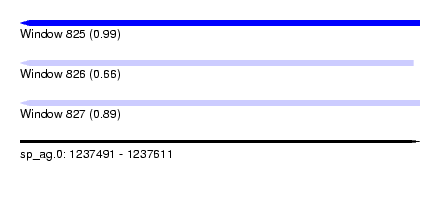
| Sequence ID | sp_ag.0 |
|---|---|
| Location | 1,237,491 – 1,237,611 |
| Length | 120 |
| Max. P | 0.985208 |

| Location | 1,237,491 – 1,237,611 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.72 |
| Mean single sequence MFE | -28.68 |
| Consensus MFE | -9.11 |
| Energy contribution | -6.86 |
| Covariance contribution | -2.25 |
| Combinations/Pair | 1.90 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.32 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
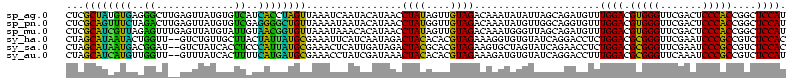
>sp_ag.0 1237491 120 - 2160267/80-200 CAGUUAAAUAUAACUGCAAAAAAUACAAAUUCUUACGCAUUAGCUGCCUAAAAAACAGCCUGCGUGAUCUUCACAAGAUUGUUUGCGUUUUGCUAGAAGGUCUUAUUUAUCAGCAAACUA .(((((....)))))((.................(((((...((((.........)))).)))))(((((((.((((((.......))))))...)))))))..........))...... ( -23.20) >sp_pn.0 773591 118 + 2038615/80-200 CAGUUAAAUAUAACUGCAAAAAAUAACACUUCUUACGCUCUAGCUGCCUAAAAACCAGCAGGCGUGACCCGAUUUGGAUUGCUCGUGUUCAA-UGACAGGUCUUAUU-AUUAGCGAGAUA ((((((....))))))..............((((((((.((.((((.........))))))))))((((.(..((((((.......))))))-...).)))).....-......)))).. ( -24.40) >sp_mu.0 891226 118 - 2030921/80-200 CAGUUAAAUAUAACUGCAAAAAAUACAAAUUCUUACGCAGUAGCUGCCUAAAAACCAGCCUGUGUGAUCAAUAACAAAUUGCUUGUGUUUGU-UGAUUGGUCUUAUU-GUUAACAAGCUA ((((((....))))))........((((......((((((..((((.........))))))))))((((((((((((((.......))))))-).)))))))...))-)).......... ( -25.90) >sy_ha.0 550673 117 - 2685015/80-200 UCAGUAAAUAUAACUGACAAAUCAAACAAUAAUUUCGCAGUAGCUGCGUAAUAG-CACUCUGCAUCGCCUA-ACAGCAUCUCCUACGUGCUGUUAACGCGAUUCAACCCU-AGUAGGAUA (((((.......)))))...(((..((.........((((.((.(((......)-))))))))(((((.((-(((((((.......)))))))))..)))))........-.))..))). ( -32.80) >sy_sa.0 543911 117 - 2516575/80-200 -CAGUUAUAAUAACUGGCAAAGAAAACAAUAAUUUCGCAGUAGCUGCGUAAUAG-CACUCUGCAUCGCCUA-ACAGCAUCUCCUAUGUGCUGUUAACGCGAUUCAACCUUUAGUAGGAUA -((((((...)))))).....(((............((((.((.(((......)-)))))))).((((.((-(((((((.......)))))))))..)))))))..(((.....)))... ( -33.30) >sy_au.0 875945 117 + 2809422/80-200 CAGUUUAUAAUAACUGGCAAAUCAAACAAUAAUUUCGCAGUAGCUGCCUAAUCG-CACUCUGCAUCGCCUA-ACAGCAUUUCCUAUGUGCUGUUAACGCGAUUCAACCUU-AAUAGGAUA (((((......)))))....................((((.((.(((......)-))))))))(((((.((-(((((((.......)))))))))..)))))....(((.-...)))... ( -32.50) >consensus CAGUUAAAUAUAACUGCAAAAAAAAACAAUAAUUACGCAGUAGCUGCCUAAAAA_CACCCUGCAUCACCUA_ACAGCAUUGCCUAUGUGCUGUUAACGCGACUCAACCAUUAGCAAGAUA ....................................((((...................)))).................((((((..........................)))))).. ( -9.11 = -6.86 + -2.25)

| Location | 1,237,491 – 1,237,609 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.49 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -21.37 |
| Energy contribution | -17.24 |
| Covariance contribution | -4.13 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.663305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 1237491 118 - 2160267/240-360 A--UAGACUCGCUAUGUGAGGGCUUGAGUUAUGUGUCAUCACCUAGUUAAAUCAAUACAUAACCUAUAGUUGUAGACAAAUAUAUUAGCAGAUGUUUGGACGUGGGUUCGACUCCCACCG .--....(((((...)))))((((.(.(((((((((..................))))))))))...))))((...(((((((........))))))).))(((((.......))))).. ( -25.67) >sp_pn.0 773591 118 + 2038615/240-360 A--UAGACUCGCAGUUUCUAGACUUGAGUUAUGUGUCGAGGGGCUGUUAAAAUAAUACAUAACCUAUGGUUGUAGACAAAUAUGUUGGCAGGUGUUUGGACGUGGGUUCGACUCCCACCG .--..((((((.((((....))))))))))..((.(((((.(.(((((((.(((....((((((...)))))).......))).))))))).).)))))))(((((.......))))).. ( -30.60) >sp_mu.0 891226 118 - 2030921/240-360 A--CUGACUCGCAUCGUUAGAGUUUGAGUUAUGUAUUGUAACGGUGUUAAAUAAACACAUAACCUAUAGUUGUAGACAAAUGGGUUAGCAGAUGUUUGGACGUGGGUUCGACUCCCACCG .--(((((.......))))).(((..((((((.....))))).(((((.....))))).((((((((..(((....)))))))))))........)..)))(((((.......))))).. ( -29.90) >sy_ha.0 550673 118 - 2685015/240-360 AAUCAAACUAGCAUAAUACUGGUU--GUCUGUUGCUUACUAUUAUGCGAAAUUCAUCAAUAGACUACACACGUAGAAAGGUGUGUAUCAGGACCUCUGGACGCGGGUUCGAAUCCCGCCG .....((((((.......))))))--(((((((((..........))((......)))))))))(((((((........)))))))((((.....))))..(((((.......))))).. ( -30.30) >sy_sa.0 543911 118 - 2516575/240-360 AAUCAAACUAGCAUAAUGACGGAU--GUCUAUCACCUCCCAUUAUGCGAAACUCAUUGAUAGACUACGCACGUAGAAGUGCUAGUAUCAGAACCUCUGGACGCGGGUUCGAAUCCCGCCG .(((((....((((((((..(((.--((.....)).)))))))))))(.....).)))))..((((.((((......)))))))).((((.....))))..(((((.......))))).. ( -34.80) >sy_au.0 875945 118 + 2809422/240-360 AAUCAAACUAGCAUCAUGUUGGUU--GUUUAUCACUUUUCAUGAUGCGAAACCUAUCGAUAAACUACACACGUAGAAAGAUGUGUAUCAGGACCUUUGGACGCGGGUUCAAAUCCCGCCG ..(((((...((((((((..(((.--.......)))...))))))))((.((.((((......((((....))))...)))).)).))......)))))..(((((.......))))).. ( -31.00) >consensus A__CAAACUAGCAUAAUGAGGGUU__AGUUAUGUCUUAUCAUGAUGCGAAAUUAAUACAUAAACUACACACGUAGAAAAAUGUGUAUCAAGACCUUUGGACGCGGGUUCGAAUCCCACCG ..........((((((((..((.............))..))))))))................((((....))))..........................(((((.......))))).. (-21.37 = -17.24 + -4.13)

| Location | 1,237,491 – 1,237,611 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.89 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -24.30 |
| Energy contribution | -20.17 |
| Covariance contribution | -4.13 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
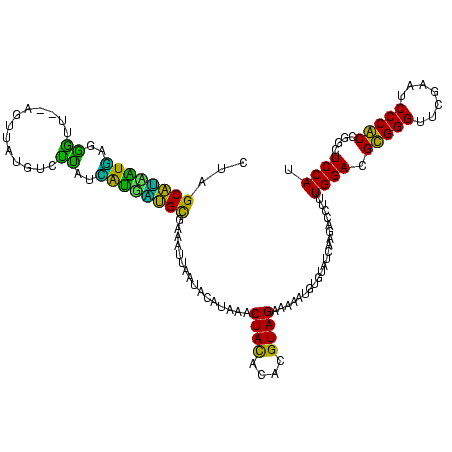
>sp_ag.0 1237491 120 - 2160267/247-367 CUCGCUAUGUGAGGGCUUGAGUUAUGUGUCAUCACCUAGUUAAAUCAAUACAUAACCUAUAGUUGUAGACAAAUAUAUUAGCAGAUGUUUGGACGUGGGUUCGACUCCCACCGGCUCCAU ........(((.(((((...(((((((((..................)))))))))........((...(((((((........))))))).))(((((.......))))).)))))))) ( -25.67) >sp_pn.0 773591 120 + 2038615/247-367 CUCGCAGUUUCUAGACUUGAGUUAUGUGUCGAGGGGCUGUUAAAAUAAUACAUAACCUAUGGUUGUAGACAAAUAUGUUGGCAGGUGUUUGGACGUGGGUUCGACUCCCACCGGCUCCAU ((((.((((....))))))))...........(((((((................((.(((.((((.((((....)))).)))).)))..))..(((((.......)))))))))))).. ( -32.40) >sp_mu.0 891226 120 - 2030921/247-367 CUCGCAUCGUUAGAGUUUGAGUUAUGUAUUGUAACGGUGUUAAAUAAACACAUAACCUAUAGUUGUAGACAAAUGGGUUAGCAGAUGUUUGGACGUGGGUUCGACUCCCACCGGCUCCAU ...(((((((((.(((...........))).))))))))).....((((((.((((((((..(((....)))))))))))...).)))))(((.(((((.......)))))....))).. ( -30.70) >sy_ha.0 550673 118 - 2685015/247-367 CUAGCAUAAUACUGGUU--GUCUGUUGCUUACUAUUAUGCGAAAUUCAUCAAUAGACUACACACGUAGAAAGGUGUGUAUCAGGACCUCUGGACGCGGGUUCGAAUCCCGCCGUCUCCAC ...((((((((..(((.--.......)))...))))))))..............((.(((((((........))))))))).(((......((((((((.......)))).))))))).. ( -33.50) >sy_sa.0 543911 118 - 2516575/247-367 CUAGCAUAAUGACGGAU--GUCUAUCACCUCCCAUUAUGCGAAACUCAUUGAUAGACUACGCACGUAGAAGUGCUAGUAUCAGAACCUCUGGACGCGGGUUCGAAUCCCGCCGUCUCCAC ...((((((((..(((.--((.....)).)))))))))))........(((((..((((.((((......))))))))))))).......(((((((((.......)))).))))).... ( -36.90) >sy_au.0 875945 118 + 2809422/247-367 CUAGCAUCAUGUUGGUU--GUUUAUCACUUUUCAUGAUGCGAAACCUAUCGAUAAACUACACACGUAGAAAGAUGUGUAUCAGGACCUUUGGACGCGGGUUCAAAUCCCGCCGUCUCCAU ...((((((((..(((.--.......)))...))))))))((.((.((((......((((....))))...)))).)).)).(((......((((((((.......)))).))))))).. ( -34.30) >consensus CUAGCAUAAUGAGGGUU__AGUUAUGUCUUAUCAUGAUGCGAAAUUAAUACAUAAACUACACACGUAGAAAAAUGUGUAUCAAGACCUUUGGACGCGGGUUCGAAUCCCACCGGCUCCAU ...((((((((..((.............))..))))))))................((((....)))).....................((((.(((((.......)))))....)))). (-24.30 = -20.17 + -4.13)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:07 2006