| Sequence ID | sp_mu.0 |
|---|---|
| Location | 722,198 – 722,316 |
| Length | 118 |
| Max. P | 0.909808 |

| Location | 722,198 – 722,315 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.35 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.30 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
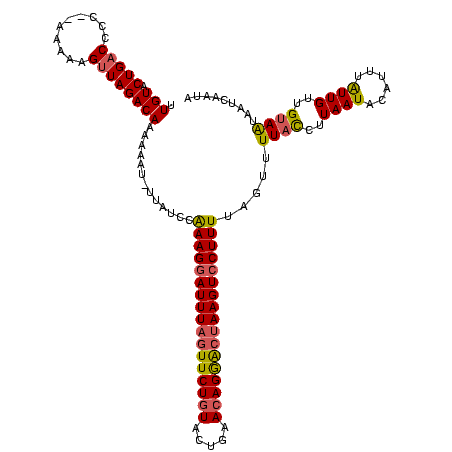
>sp_mu.0 722198 117 - 2030921/0-120 GUGUACUGACCCC--AAAAAGUUAGACAAAAAAU-UUAAGUAAAGGAUUUCGUUCUGUACUGUACAGGACUAAGUCCUUUUAGUUUUACCUUAAUACGUUUGUUGUUGUAAUAAUCAAUA .....(((((...--.....)))))(((((....-(((((.(((((((((.((((((((...)))))))).))))))))).........)))))....)))))(((((.......))))) ( -26.10) >sp_ag.0 893897 118 - 2160267/0-120 UUGUACUGACCCC--AAAAAGUUAGACAAAAAAUAUUAUUGAAAGGAUUUAAUUCUAUACUGAACAGGACUAAGUCCUUUUAGUUUUUCUUUAAUGCAUUUAUUGUUGUAGUAAAAAAUA ((((.(((((...--.....)))))))))........((((((((((((((.((((.........)))).)))))))))))))).....((((.((((........)))).))))..... ( -24.00) >sp_pn.0 876248 118 + 2038615/0-120 UUGUACUGCCCCCCAAAAAAGUUAGACAAUUAAU-UUAUCCGAAG-AUUUAGUUCUGUAUUGCACAGAGCUAAGUCCUUUUAGUUUUAUCUUAAUUCUCUUAUUGUUGUAAUAAUCAAUA .............(((...(((.(((.((((((.-.((.(..(((-((((((((((((.....)))))))))))))..))..)...))..)))))).))).))).)))............ ( -21.80) >consensus UUGUACUGACCCC__AAAAAGUUAGACAAAAAAU_UUAUCCAAAGGAUUUAGUUCUGUACUGAACAGGACUAAGUCCUUUUAGUUUUACCUUAAUACAUUUAUUGUUGUAAUAAUCAAUA .(((.(((((..........)))))))).............(((((((((((((((((.....))))))))))))))))).....((((..((((......))))..))))......... (-19.62 = -20.30 + 0.68)

| Location | 722,198 – 722,316 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.73 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -19.07 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
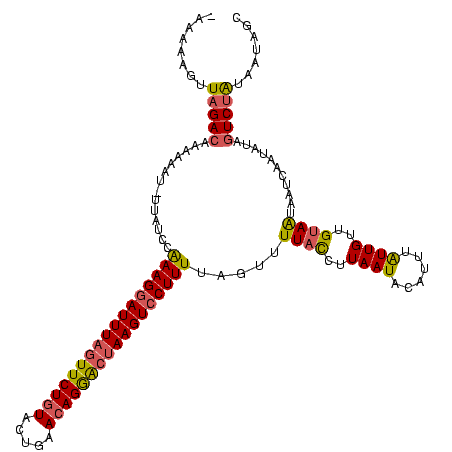
>sp_mu.0 722198 118 - 2030921/14-134 -AAAAAGUUAGACAAAAAAU-UUAAGUAAAGGAUUUCGUUCUGUACUGUACAGGACUAAGUCCUUUUAGUUUUACCUUAAUACGUUUGUUGUUGUAAUAAUCAAUAUAGUCUACAAUAGC -.....((.((((..(((((-(.....(((((((((.((((((((...)))))))).))))))))).))))))................(((((.......)))))..))))))...... ( -26.00) >sp_ag.0 893897 119 - 2160267/14-134 -AAAAAGUUAGACAAAAAAUAUUAUUGAAAGGAUUUAAUUCUAUACUGAACAGGACUAAGUCCUUUUAGUUUUUCUUUAAUGCAUUUAUUGUUGUAGUAAAAAAUAUACUCGGUGAUAGC -.....((((((.((((......((((((((((((((.((((.........)))).)))))))))))))))))).))))))....((((..(((.((((.......)))))))..)))). ( -23.00) >sp_pn.0 876248 118 + 2038615/14-134 AAAAAAGUUAGACAAUUAAU-UUAUCCGAAG-AUUUAGUUCUGUAUUGCACAGAGCUAAGUCCUUUUAGUUUUAUCUUAAUUCUCUUAUUGUUGUAAUAAUCAAUAUAGUCUAUAAUGGC ........(((((((((((.-.((.(..(((-((((((((((((.....)))))))))))))..))..)...))..)))))).......(((((.......)))))..)))))....... ( -23.60) >consensus _AAAAAGUUAGACAAAAAAU_UUAUCCAAAGGAUUUAGUUCUGUACUGAACAGGACUAAGUCCUUUUAGUUUUACCUUAAUACAUUUAUUGUUGUAAUAAUCAAUAUAGUCUAUAAUAGC ........(((((..............(((((((((((((((((.....))))))))))))))))).....((((..((((......))))..))))...........)))))....... (-19.07 = -19.87 + 0.79)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:04 2006