| Sequence ID | sy_ha.0 |
|---|---|
| Location | 95 – 1,232,408 |
| Length | 1232313 |
| Max. P | 0.929297 |

| Location | 1,232,332 – 1,232,408 |
|---|---|
| Length | 76 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 62.24 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -10.08 |
| Energy contribution | -9.84 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
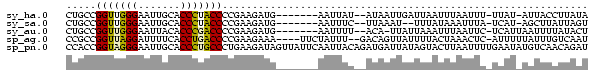
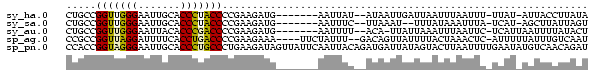
>sy_ha.0 1232332 76 - 2685015 CUGCCGGUUGGGAAUUGCACCCUACCCCGAAGAUG-------AAUUAU--AUAAUUGAUUAAUUUAAUUU-UUAU-AUUACCUUAUA .....(((.(((.......))).)))..(((((((-------(((((.--.........)))))).))))-))..-........... ( -10.30) >sy_sa.0 1211975 74 - 2516575 CUGCCGGUUGGGAAUUGCACCCUACCCCGAAGAUG-------AAUUUC--UUAAAU--UUUAUAAAUUUA-UCAU-AGCUUAUUAGU ..((.(((.(((.......))).))).....((((-------(((((.--(.....--...).)))))))-))..-.))........ ( -13.30) >sy_au.0 1565451 76 + 2809422 CUGCCGGUUGGGAAUUACACCCGACCCCGAAGAUG-------AAUUUU--ACA-UUAUUAAAUUUAAUUC-UCAUUAAUUUUAUACU .....(((((((.......)))))))..(((..((-------(((((.--...-.....))))))).)))-................ ( -13.90) >sp_ag.0 1420944 80 - 2160267 CCGCCGGUUAGGAUUUUCACCUGACCCCGAAGAAA----UUCUAUUU--GACAGUUAUUUUACUAAACUC-AUUUUUAUUUGUCAAU .....(((((((.......)))))))..(((....----)))...((--(((((................-........))))))). ( -16.06) >sp_pn.0 173968 87 + 2038615 CCACCGGUAGGGAAUUGCACCCUGCCCUGAAGAUAGUUAUUCAAUUACAGAUGAUUAUAGUACUUAAUUUUGAAUAUGUCAACAGAU .....(((((((.......)))))))(((..((((..(((((((........(((((.......))))))))))))))))..))).. ( -20.70) >consensus CUGCCGGUUGGGAAUUGCACCCUACCCCGAAGAUG_______AAUUUC__AUAAUUAUUUUAUUUAAUUC_UCAU_AAUUUAUUAAU .....(((((((.......)))))))............................................................. (-10.08 = -9.84 + -0.24)
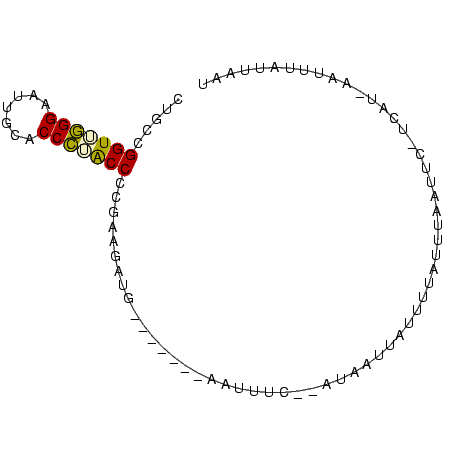
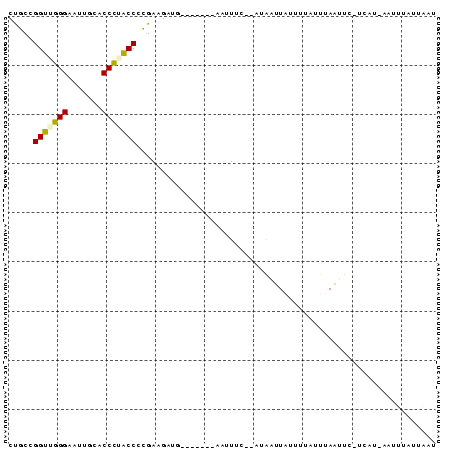
| Location | 1,232,332 – 1,232,408 |
|---|---|
| Length | 76 |
| Sequences | 5 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 62.24 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -10.08 |
| Energy contribution | -9.84 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1232332 76 - 2685015/0-87 CUGCCGGUUGGGAAUUGCACCCUACCCCGAAGAUG-------AAUUAU--AUAAUUGAUUAAUUUAAUUU-UUAU-AUUACCUUAUA .....(((.(((.......))).)))..(((((((-------(((((.--.........)))))).))))-))..-........... ( -10.30) >sy_sa.0 1211975 74 - 2516575/0-87 CUGCCGGUUGGGAAUUGCACCCUACCCCGAAGAUG-------AAUUUC--UUAAAU--UUUAUAAAUUUA-UCAU-AGCUUAUUAGU ..((.(((.(((.......))).))).....((((-------(((((.--(.....--...).)))))))-))..-.))........ ( -13.30) >sy_au.0 1565451 76 + 2809422/0-87 CUGCCGGUUGGGAAUUACACCCGACCCCGAAGAUG-------AAUUUU--ACA-UUAUUAAAUUUAAUUC-UCAUUAAUUUUAUACU .....(((((((.......)))))))..(((..((-------(((((.--...-.....))))))).)))-................ ( -13.90) >sp_ag.0 1420944 80 - 2160267/0-87 CCGCCGGUUAGGAUUUUCACCUGACCCCGAAGAAA----UUCUAUUU--GACAGUUAUUUUACUAAACUC-AUUUUUAUUUGUCAAU .....(((((((.......)))))))..(((....----)))...((--(((((................-........))))))). ( -16.06) >sp_pn.0 173968 87 + 2038615/0-87 CCACCGGUAGGGAAUUGCACCCUGCCCUGAAGAUAGUUAUUCAAUUACAGAUGAUUAUAGUACUUAAUUUUGAAUAUGUCAACAGAU .....(((((((.......)))))))(((..((((..(((((((........(((((.......))))))))))))))))..))).. ( -20.70) >consensus CUGCCGGUUGGGAAUUGCACCCUACCCCGAAGAUG_______AAUUUC__AUAAUUAUUUUAUUUAAUUC_UCAU_AAUUUAUUAAU .....(((((((.......)))))))............................................................. (-10.08 = -9.84 + -0.24)
| Location | 95 – 214 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -27.13 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
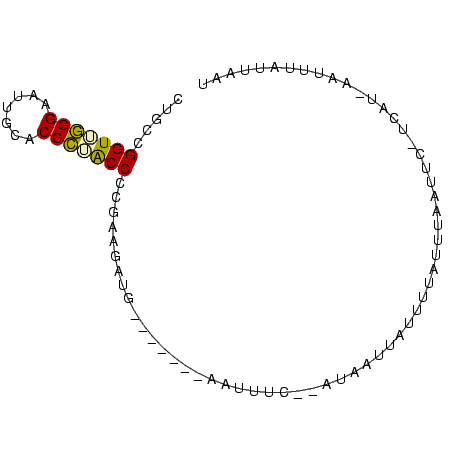
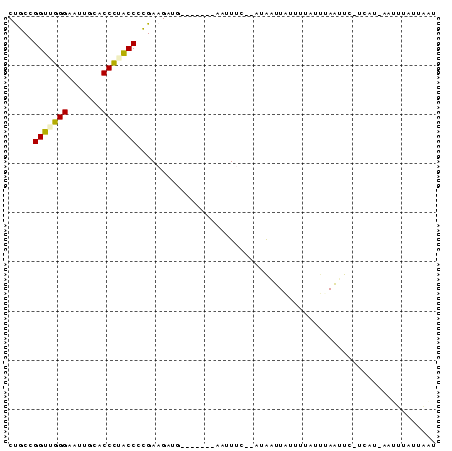
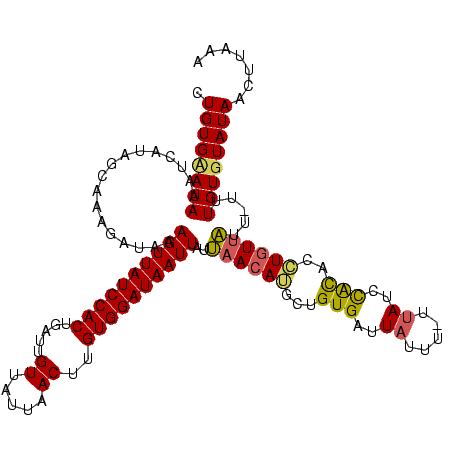
>sy_ha.0 95 119 + 2685015/280-400 GUGUGAAUAAAUCAUAGCAUAUAUAAAGUUAUCCACUGAAUGUUAUUAACUUGUGGAUAAUUAUUAACAUGCUGUGAUUAUUU-UUAUCCACUUGUCUAAAUGUCUGUGUAUAAGUCCUA (..((((..(((((((((((......((((((((((................))))))))))......)))))))))))...)-)))..)(((((((...........).)))))).... ( -27.69) >sy_au.0 123 119 + 2809422/280-400 CUGUGCAUAACUAAUAAGCAAGAUAAAGUUAUCCACCGAUUGUUAUUAACUUGUGGAUAAUUAUUAACAUGGUGUGUUUAGAAGUUAUCCACGGCUGUUAUU-UUUGUGUAUAACUUAAA .(((((((((.(((((.((......((((((...((.....))...))))))((((((((((...(((((...)))))....)))))))))).)))))))..-.)))))))))....... ( -27.60) >sy_sa.0 85 118 + 2516575/280-400 CUGUGUAUAAAUCACAGUUAGGUAAUAGUUAUCCACUGAUUGUGCGUAACUUGUGGAUAAUUAUCAACAGCCUGUGAUUCUUU-UUAUUUGUACCUGUUGUU-UUUGUGUAUAACUUUAA ....(((((((((((((....((.((((((((((((.....((.....))..))))))))))))..))...))))))).....-....))))))..(((((.-.......)))))..... ( -26.10) >consensus CUGUGAAUAAAUCAUAGCAAAGAUAAAGUUAUCCACUGAUUGUUAUUAACUUGUGGAUAAUUAUUAACAUGCUGUGAUUAUUU_UUAUCCACACCUGUUAUU_UUUGUGUAUAACUUAAA .((((((((.................((((((((((.....((.....))..))))))))))..((((((...(((..((.....))..)))..)))))).....))))))))....... (-14.30 = -14.53 + 0.23)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:37 2006