
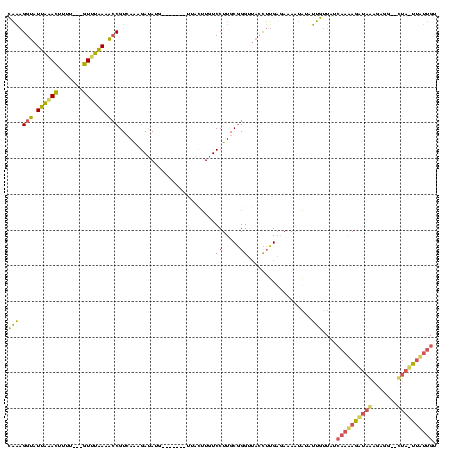
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 660,473 – 888,822 |
| Length | 228349 |
| Max. P | 0.962234 |

| Location | 888,707 – 888,822 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 54.57 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -9.39 |
| Energy contribution | -9.30 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 888707 115 - 2030921/0-120 CAGAGGUAUUAGACCAA----GUCUAAGACCGUCAGAGAUAUGCGUCUAGUUGUUUGUCCUUGCGGGAUACCUUGUGAAA-UAUAUUGUUAUCAAGAGAUAAAGAGGGUUUAUUUCUUGU ..((((((((((((...----))))....((((.((.((((.(((......))).)))))).))))))))))))......-...........(((((((((((.....))))))))))). ( -32.00) >sp_ag.0 1006131 108 - 2160267/0-120 CAAAGGUAUCAAACUUUU---GUUUGACACCGUCAAAGAGAUA-------UUUGUUGUCCUUGCGGGUUACCUUGAGAAAAUAUAUUGUUAUCAAAAGCUAGCUCGG--CUAGCUUUUGU ..((((((((((((....---))))))..((((..(((.((((-------.....)))))))))))..))))))..................(((((((((((...)--)))))))))). ( -34.40) >sp_pn.0 759602 110 + 2038615/0-120 UUAGGGUAUUUUAUUUUUAUAAUUGAAAGACGUGAAUGAUAUG--------AACAUGUCCUUGCUGGUGCUUAGGAAAAAAAUUAUAAGUAUGUCAAGUUUAAGAAAAACU--UGAUUGU ....((((......(((((....)))))((((((.........--------..))))))..)))).(((((((.((......)).)))))))(((((((((.....)))))--))))... ( -18.90) >consensus CAAAGGUAUUAAACUUUU___GUUUAAAACCGUCAAAGAUAUG_______UUACUUGUCCUUGCGGGUUACCUUGAGAAAAUAUAUUGUUAUCAAAAGAUAAAGAGG__CUA_UUAUUGU (((.(((.((((((.......)))))).)))...........................((.....))..................)))....((((((((((.......)))))))))). ( -9.39 = -9.30 + -0.09)

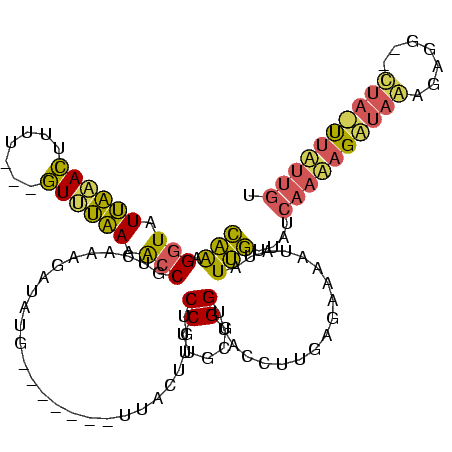
| Location | 888,707 – 888,822 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 54.57 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -4.91 |
| Energy contribution | -5.47 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.27 |
| Mean z-score | -3.85 |
| Structure conservation index | 0.20 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 888707 115 - 2030921/0-120 ACAAGAAAUAAACCCUCUUUAUCUCUUGAUAACAAUAUA-UUUCACAAGGUAUCCCGCAAGGACAAACAACUAGACGCAUAUCUCUGACGGUCUUAGAC----UUGGUCUAAUACCUCUG .(((((.(((((.....))))).)))))...........-.......((((((.((....)).........(((((.((..(((..(.....)..))).----.)))))))))))))... ( -24.40) >sp_ag.0 1006131 108 - 2160267/0-120 ACAAAAGCUAG--CCGAGCUAGCUUUUGAUAACAAUAUAUUUUCUCAAGGUAACCCGCAAGGACAACAAA-------UAUCUCUUUGACGGUGUCAAAC---AAAAGUUUGAUACCUUUG .((((((((((--(...)))))))))))................((((((....((....))........-------.....)))))).((((((((((---....)))))))))).... ( -34.63) >sp_pn.0 759602 110 + 2038615/0-120 ACAAUCA--AGUUUUUCUUAAACUUGACAUACUUAUAAUUUUUUUCCUAAGCACCAGCAAGGACAUGUU--------CAUAUCAUUCACGUCUUUCAAUUAUAAAAAUAAAAUACCCUAA ....(((--(((((.....)))))))).....((((((((..........((....))((((((.((..--------.........)).)))))).))))))))................ ( -15.40) >consensus ACAAAAA_UAG__CCUCUUUAACUUUUGAUAACAAUAUAUUUUCUCAAGGUAACCCGCAAGGACAAAAAA_______CAUAUCUUUGACGGUCUUAAAC___AAAAGUAUAAUACCUUUG ......................................................((....))...........................(((.((((((.......)))))).))).... ( -4.91 = -5.47 + 0.56) # Strand winner: forward (0.64)


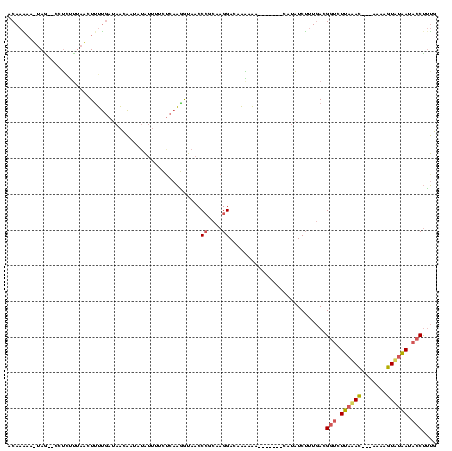
| Location | 660,473 – 660,551 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -17.33 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 660473 78 + 2030921 UUCUAUUUUAAGUGAGAAAAGCAGAAGUGAGAACUUCUCGCCUUGCGACUUAAAGUUGUCUGGCCCAACAUGUUAAAG ......((((((((......(.((((((....)))))))(((..(((((.....)))))..)))....))).))))). ( -18.20) >sp_ag.0 766044 77 - 2160267 UACUAGUUGCAAUGAGAAAAGCAGAAGUGAGAACUUCUCGCCUUGCGACUUAG-GUUGUCUGGCCCAACAUGUCAAGA .....((((...........(.((((((....)))))))(((..(((((....-)))))..))).))))......... ( -19.70) >sp_pn.0 855158 78 + 2038615 UCAUAUAAAAGUUGAGAAAAGCAGAAGUGAGAGCUUCUCGCCUUGUGACAUUAAGUUGCCUGGCCCUACGGAUGAAAA ((((......(((......)))((((((....)))))).(((..(..((.....))..)..))).......))))... ( -19.40) >consensus UACUAUUUAAAAUGAGAAAAGCAGAAGUGAGAACUUCUCGCCUUGCGACUUAAAGUUGUCUGGCCCAACAUGUCAAAA ...........(((......(.((((((....)))))))(((..(((((.....)))))..)))....)))....... (-17.33 = -16.73 + -0.60)
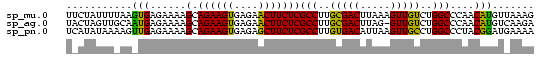
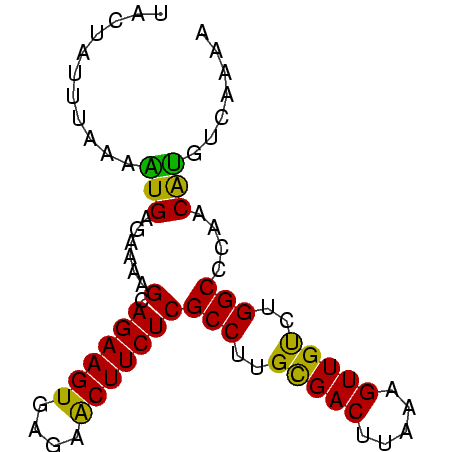
| Location | 660,473 – 660,551 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 78.63 |
| Mean single sequence MFE | -19.10 |
| Consensus MFE | -17.33 |
| Energy contribution | -16.73 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 660473 78 + 2030921/0-78 UUCUAUUUUAAGUGAGAAAAGCAGAAGUGAGAACUUCUCGCCUUGCGACUUAAAGUUGUCUGGCCCAACAUGUUAAAG ......((((((((......(.((((((....)))))))(((..(((((.....)))))..)))....))).))))). ( -18.20) >sp_ag.0 766044 77 - 2160267/0-78 UACUAGUUGCAAUGAGAAAAGCAGAAGUGAGAACUUCUCGCCUUGCGACUUAG-GUUGUCUGGCCCAACAUGUCAAGA .....((((...........(.((((((....)))))))(((..(((((....-)))))..))).))))......... ( -19.70) >sp_pn.0 855158 78 + 2038615/0-78 UCAUAUAAAAGUUGAGAAAAGCAGAAGUGAGAGCUUCUCGCCUUGUGACAUUAAGUUGCCUGGCCCUACGGAUGAAAA ((((......(((......)))((((((....)))))).(((..(..((.....))..)..))).......))))... ( -19.40) >consensus UACUAUUUAAAAUGAGAAAAGCAGAAGUGAGAACUUCUCGCCUUGCGACUUAAAGUUGUCUGGCCCAACAUGUCAAAA ...........(((......(.((((((....)))))))(((..(((((.....)))))..)))....)))....... (-17.33 = -16.73 + -0.60)
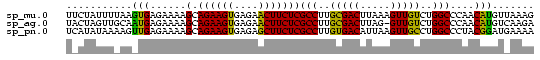
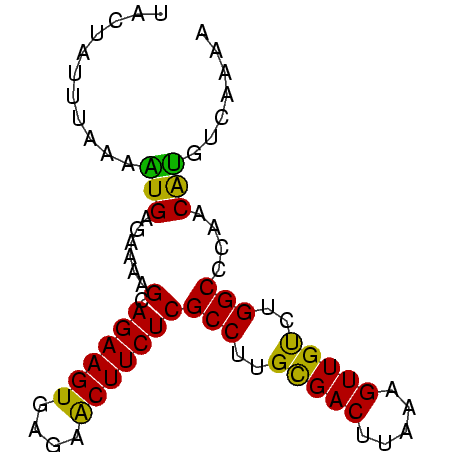
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:03 2006