| Sequence ID | sp_mu.0 |
|---|---|
| Location | 642,196 – 642,315 |
| Length | 119 |
| Max. P | 0.999999 |

| Location | 642,196 – 642,315 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.13 |
| Mean single sequence MFE | -31.43 |
| Consensus MFE | -17.51 |
| Energy contribution | -14.97 |
| Covariance contribution | -2.54 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.56 |
| SVM decision value | 4.73 |
| SVM RNA-class probability | 0.999944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 642196 119 - 2030921/0-120 UAUCUUUCUAAAUUGAACACGGGGCUAAUUUCCUAUGAAAAAAGAUAAUCUUUCCUAGACGCAAGAGUCUUUGU-CAGAUUUCCUAUUUUUCUACGGAAAUUUUCAGCAAGCUGAAUCGC .....(((......)))..(((....(((((((...((((((((..(((((..(..((((......))))..).-.)))))..)).))))))...)))))))(((((....)))))))). ( -25.40) >sp_ag.0 43257 119 + 2160267/0-120 UAGUUUGCCUAACUGAACACGGGAUUAAAUUUCUAGGAAAAAAGAUAAAACGACUUAGACGCAAGCGUCGUUGU-UCGUUUUCCUAUUUUUCUACGAAAUUUUUCGGCAAGCCGAAUCAU ..(((((((...(((....)))((..(((((((((((((((.((..((((((((...((((....))))...).-))))))).)).)))))))).))))))).)))))))))........ ( -36.30) >sp_pn.0 515516 120 + 2038615/0-120 UAUCUUGAUGACUUGAAUUUUGGUCUCAUUUCUUAGGAAAAAAGAUAAGCUUCCUAGCACUCGUCGAGUGCGACGUCACCUUCCUAUUUUUCUACAGAAAUGUUCGGCAAGCCGAACCGU ......((.((((........))))))((((((((((((((.((....(((....)))...(((((....)))))........)).)))))))).))))))((((((....))))))... ( -32.60) >consensus UAUCUUGCUAAAUUGAACACGGGACUAAUUUCCUAGGAAAAAAGAUAAACUUACUUAGACGCAAGAGUCGUUGU_UCGAUUUCCUAUUUUUCUACGGAAAUUUUCGGCAAGCCGAAUCGU .....................((...(((((((((((((((.((...............(....)..(((......)))....)).)))))))).)))))))(((((....))))))).. (-17.51 = -14.97 + -2.54)


| Location | 642,196 – 642,315 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.13 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -20.39 |
| Energy contribution | -17.96 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.85 |
| SVM RNA-class probability | 0.999956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 642196 119 - 2030921/0-120 GCGAUUCAGCUUGCUGAAAAUUUCCGUAGAAAAAUAGGAAAUCUG-ACAAAGACUCUUGCGUCUAGGAAAGAUUAUCUUUUUUCAUAGGAAAUUAGCCCCGUGUUCAAUUUAGAAAGAUA (((.(((((....)))))(((((((...((((((.(((.(((((.-.(..((((......))))..)..))))).)))))))))...))))))).....))).................. ( -28.00) >sp_ag.0 43257 119 + 2160267/0-120 AUGAUUCGGCUUGCCGAAAAAUUUCGUAGAAAAAUAGGAAAACGA-ACAACGACGCUUGCGUCUAAGUCGUUUUAUCUUUUUUCCUAGAAAUUUAAUCCCGUGUUCAGUUAGGCAAACUA .((((((((....)))))(((((((...((((((.((((((((((-.....((((....))))....))))))).)))))))))...)))))))..........)))............. ( -29.10) >sp_pn.0 515516 120 + 2038615/0-120 ACGGUUCGGCUUGCCGAACAUUUCUGUAGAAAAAUAGGAAGGUGACGUCGCACUCGACGAGUGCUAGGAAGCUUAUCUUUUUUCCUAAGAAAUGAGACCAAAAUUCAAGUCAUCAAGAUA ...((((((....)))))).(((((((......)))))))((((((...((((((...))))))..((...(((((.((((.....)))).))))).)).........))))))...... ( -35.80) >consensus ACGAUUCGGCUUGCCGAAAAUUUCCGUAGAAAAAUAGGAAAACGA_ACAACGACCCUUGCGUCUAAGAAAGAUUAUCUUUUUUCCUAGGAAAUUAGACCCGUGUUCAAUUAAGCAAGAUA ....(((((....)))))(((((((...((((((.(((.((((........(((......))).......)))).)))))))))...))))))).......................... (-20.39 = -17.96 + -2.43) # Strand winner: reverse (0.75)

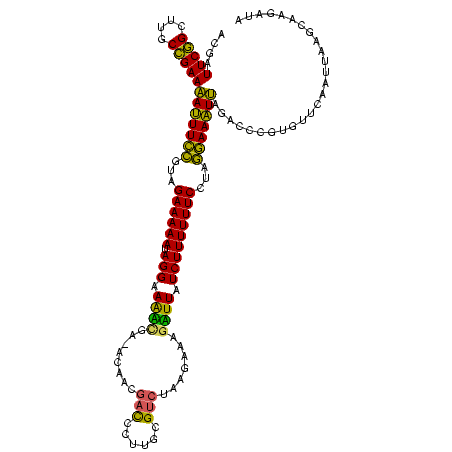
| Location | 642,196 – 642,315 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.13 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -20.72 |
| Energy contribution | -18.07 |
| Covariance contribution | -2.65 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 6.94 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 642196 119 - 2030921/2-122 UCUUUCUAAAUUGAACACGGGGCUAAUUUCCUAUGAAAAAAGAUAAUCUUUCCUAGACGCAAGAGUCUUUGU-CAGAUUUCCUAUUUUUCUACGGAAAUUUUCAGCAAGCUGAAUCGCCC ...(((......)))....((((.(((((((...((((((((..(((((..(..((((......))))..).-.)))))..)).))))))...)))))))(((((....)))))..)))) ( -31.50) >sp_ag.0 43257 119 + 2160267/2-122 GUUUGCCUAACUGAACACGGGAUUAAAUUUCUAGGAAAAAAGAUAAAACGACUUAGACGCAAGCGUCGUUGU-UCGUUUUCCUAUUUUUCUACGAAAUUUUUCGGCAAGCCGAAUCAUCC (((((((...(((....)))((..(((((((((((((((.((..((((((((...((((....))))...).-))))))).)).)))))))).))))))).))))))))).......... ( -36.10) >sp_pn.0 515516 120 + 2038615/2-122 UCUUGAUGACUUGAAUUUUGGUCUCAUUUCUUAGGAAAAAAGAUAAGCUUCCUAGCACUCGUCGAGUGCGACGUCACCUUCCUAUUUUUCUACAGAAAUGUUCGGCAAGCCGAACCGUCC ....((.((((........))))))((((((((((((((.((....(((....)))...(((((....)))))........)).)))))))).))))))((((((....))))))..... ( -32.60) >consensus UCUUGCUAAAUUGAACACGGGACUAAUUUCCUAGGAAAAAAGAUAAACUUACUUAGACGCAAGAGUCGUUGU_UCGAUUUCCUAUUUUUCUACGGAAAUUUUCGGCAAGCCGAAUCGUCC ...................((((.(((((((((((((((.((...............(....)..(((......)))....)).)))))))).)))))))(((((....)))))..)))) (-20.72 = -18.07 + -2.65)

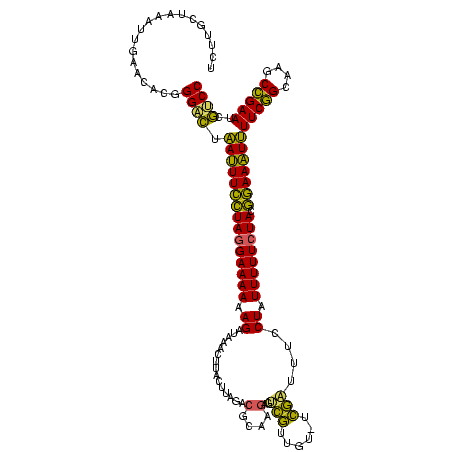
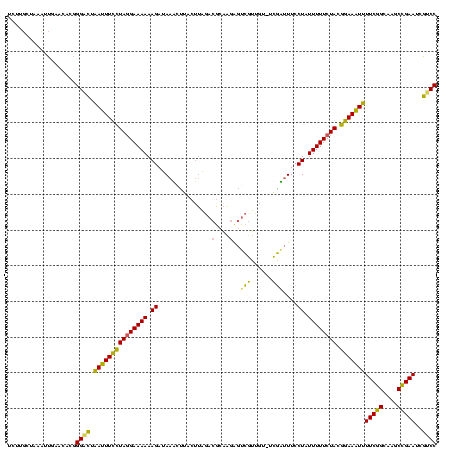
| Location | 642,196 – 642,315 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.13 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -25.36 |
| Energy contribution | -22.59 |
| Covariance contribution | -2.76 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.74 |
| SVM decision value | 6.69 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 642196 119 - 2030921/2-122 GGGCGAUUCAGCUUGCUGAAAAUUUCCGUAGAAAAAUAGGAAAUCUG-ACAAAGACUCUUGCGUCUAGGAAAGAUUAUCUUUUUUCAUAGGAAAUUAGCCCCGUGUUCAAUUUAGAAAGA ((((..(((((....)))))(((((((...((((((.(((.(((((.-.(..((((......))))..)..))))).)))))))))...))))))).))))................... ( -34.20) >sp_ag.0 43257 119 + 2160267/2-122 GGAUGAUUCGGCUUGCCGAAAAAUUUCGUAGAAAAAUAGGAAAACGA-ACAACGACGCUUGCGUCUAAGUCGUUUUAUCUUUUUUCCUAGAAAUUUAAUCCCGUGUUCAGUUAGGCAAAC ((((..(((((....)))))(((((((...((((((.((((((((((-.....((((....))))....))))))).)))))))))...))))))).))))..((((......))))... ( -32.20) >sp_pn.0 515516 120 + 2038615/2-122 GGACGGUUCGGCUUGCCGAACAUUUCUGUAGAAAAAUAGGAAGGUGACGUCGCACUCGACGAGUGCUAGGAAGCUUAUCUUUUUUCCUAAGAAAUGAGACCAAAAUUCAAGUCAUCAAGA .....((((((....)))))).(((((((......)))))))((((((...((((((...))))))..((...(((((.((((.....)))).))))).)).........)))))).... ( -35.80) >consensus GGACGAUUCGGCUUGCCGAAAAUUUCCGUAGAAAAAUAGGAAAACGA_ACAACGACCCUUGCGUCUAAGAAAGAUUAUCUUUUUUCCUAGGAAAUUAGACCCGUGUUCAAUUAAGCAAGA ((((..(((((....)))))(((((((...((((((.(((.((((........(((......))).......)))).)))))))))...))))))).))))................... (-25.36 = -22.59 + -2.76) # Strand winner: reverse (0.92)

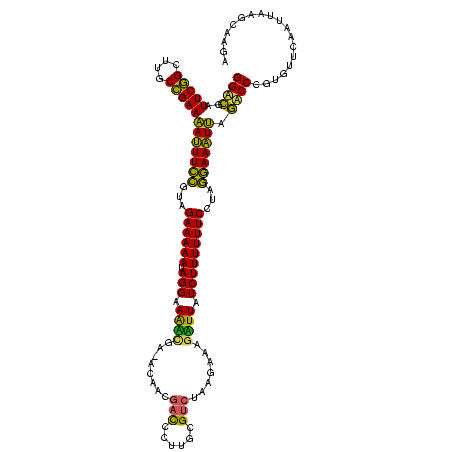

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:02:00 2006