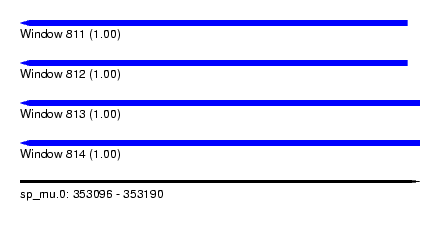
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 353,096 – 353,190 |
| Length | 94 |
| Max. P | 0.998633 |

| Location | 353,096 – 353,187 |
|---|---|
| Length | 91 |
| Sequences | 2 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.04 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -31.15 |
| Energy contribution | -32.90 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.57 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 353096 91 - 2030921/120-240 AAAAUGAGUUGCGGGUCUAGCC---CGAAACUGGGUGGUACCGCGGAGAUCAUAGAAUGACAUUCGUCCCUGUC--AAUUGGCAGGGACGGUUUUU ...((((.((((((..(((.((---((....)))))))..))))))...))))..........(((((((((((--....)))))))))))..... ( -39.10) >sp_ag.0 456464 93 + 2160267/120-240 AAAACGAGUUGCGGAUGUUAUCAUCCGAAUUUGGGUGGUACCGCGGAAAUCGUAACACGAAAUUCGUCCCUGUCAGAAUUGACAGGGA---UUUUU ...((((.((((((....(((((((((....)))))))))))))))...))))............(((((((((......))))))))---).... ( -36.60) >consensus AAAACGAGUUGCGGAUCUAACC___CGAAACUGGGUGGUACCGCGGAAAUCAUAAAACGAAAUUCGUCCCUGUC__AAUUGACAGGGA___UUUUU ...((((.((((((....(((((((((....)))))))))))))))...))))...........((((((((((......))))))))))...... (-31.15 = -32.90 + 1.75)

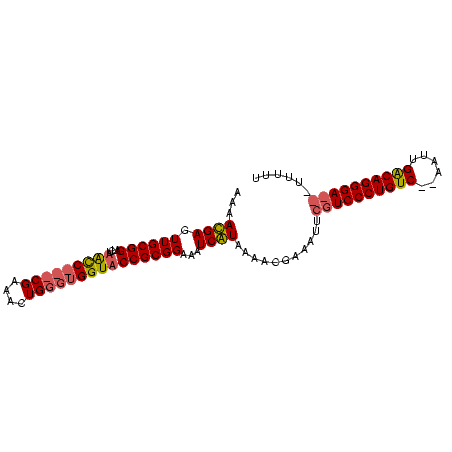
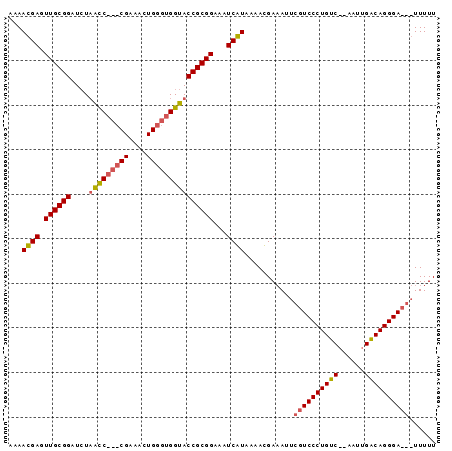
| Location | 353,096 – 353,187 |
|---|---|
| Length | 91 |
| Sequences | 2 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 76.04 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.47 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 353096 91 - 2030921/120-240 AAAAACCGUCCCUGCCAAUU--GACAGGGACGAAUGUCAUUCUAUGAUCUCCGCGGUACCACCCAGUUUCG---GGCUAGACCCGCAACUCAUUUU ......((((((((.(....--).))))))))...(((((...)))))....((((..(..(((......)---))...)..)))).......... ( -26.30) >sp_ag.0 456464 93 + 2160267/120-240 AAAAA---UCCCUGUCAAUUCUGACAGGGACGAAUUUCGUGUUACGAUUUCCGCGGUACCACCCAAAUUCGGAUGAUAACAUCCGCAACUCGUUUU .....---(((((((((....)))))))))(((...(((.....))).....((((......))......(((((....)))))))...))).... ( -28.30) >consensus AAAAA___UCCCUGCCAAUU__GACAGGGACGAAUGUCAUGCUACGAUCUCCGCGGUACCACCCAAAUUCG___GACAACACCCGCAACUCAUUUU ......((((((((((......))))))))))....(((.....))).....((((..........................)))).......... (-18.47 = -19.47 + 1.00) # Strand winner: forward (1.00)

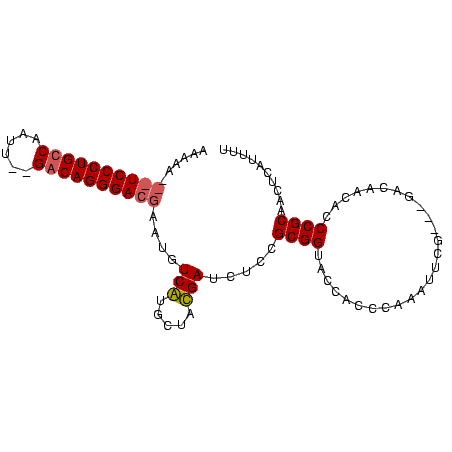
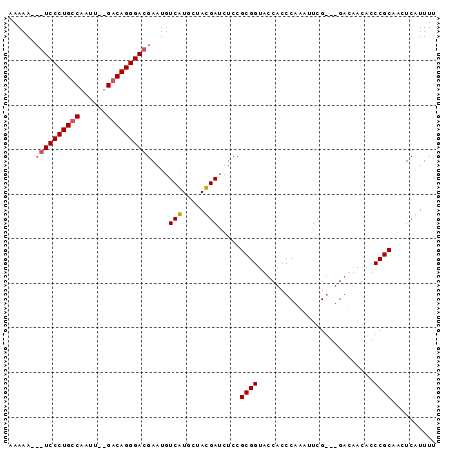
| Location | 353,096 – 353,190 |
|---|---|
| Length | 94 |
| Sequences | 2 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -39.95 |
| Consensus MFE | -31.75 |
| Energy contribution | -34.25 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.82 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 353096 94 - 2030921/124-244 AAAUGAGUUGCGGGUCUAGCC---CGAAACUGGGUGGUACCGCGGAGAUCAUAGAAUGACAUUCGUCCCUGUC--AAUUGGCAGGGACGGUUUUUGGUA ..((((.((((((..(((.((---((....)))))))..))))))...)))).......((.(((((((((((--....)))))))))))....))... ( -39.40) >sp_ag.0 456464 96 + 2160267/124-244 AAACGAGUUGCGGAUGUUAUCAUCCGAAUUUGGGUGGUACCGCGGAAAUCGUAACACGAAAUUCGUCCCUGUCAGAAUUGACAGGGA---UUUUUUGUA ..((((.((((((....(((((((((....)))))))))))))))...))))...((((((...(((((((((......))))))))---).)))))). ( -40.50) >consensus AAACGAGUUGCGGAUCUAACC___CGAAACUGGGUGGUACCGCGGAAAUCAUAAAACGAAAUUCGUCCCUGUC__AAUUGACAGGGA___UUUUUGGUA ..((((.((((((....(((((((((....)))))))))))))))...))))...((((((..((((((((((......))))))))))...)))))). (-31.75 = -34.25 + 2.50)

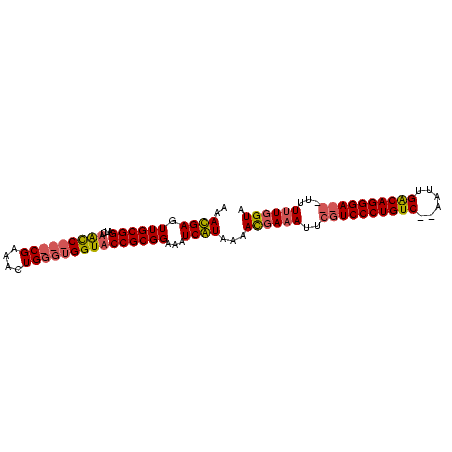
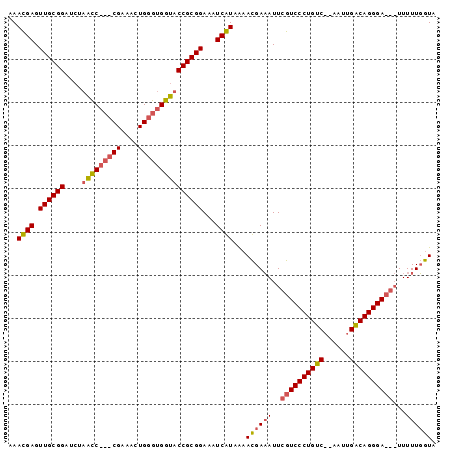
| Location | 353,096 – 353,190 |
|---|---|
| Length | 94 |
| Sequences | 2 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 75.76 |
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.47 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 353096 94 - 2030921/124-244 UACCAAAAACCGUCCCUGCCAAUU--GACAGGGACGAAUGUCAUUCUAUGAUCUCCGCGGUACCACCCAGUUUCG---GGCUAGACCCGCAACUCAUUU ((((......((((((((.(....--).))))))))...(((((...)))))......))))......((((.((---((.....)))).))))..... ( -28.30) >sp_ag.0 456464 96 + 2160267/124-244 UACAAAAAA---UCCCUGUCAAUUCUGACAGGGACGAAUUUCGUGUUACGAUUUCCGCGGUACCACCCAAAUUCGGAUGAUAACAUCCGCAACUCGUUU .........---(((((((((....)))))))))(((...(((.....))).....((((......))......(((((....)))))))...)))... ( -28.30) >consensus UACAAAAAA___UCCCUGCCAAUU__GACAGGGACGAAUGUCAUGCUACGAUCUCCGCGGUACCACCCAAAUUCG___GACAACACCCGCAACUCAUUU ..........((((((((((......))))))))))....(((.....))).....((((..........................))))......... (-18.47 = -19.47 + 1.00) # Strand winner: forward (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:56 2006