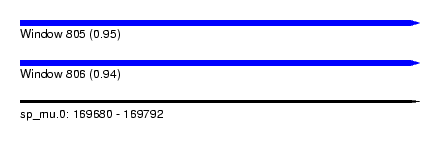
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 169,680 – 169,792 |
| Length | 112 |
| Max. P | 0.947405 |

| Location | 169,680 – 169,792 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -30.97 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
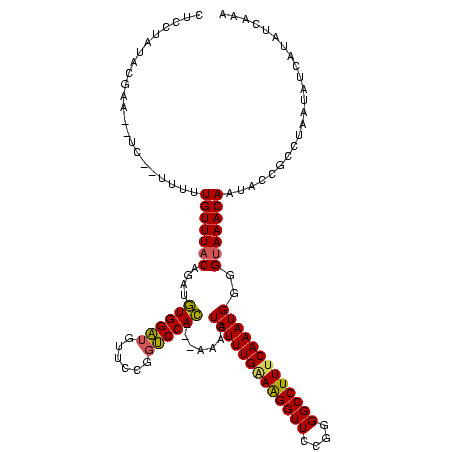
>sp_mu.0 169680 112 + 2030921/0-120 CUCCUAUACGAA--UC--UUUAUGUUUACA-AUGUGGAUGUUCCGGUCCAC---AAAUUAUUUGAAGGGUUCCGGGGCCUUUCAAAUGGGGCAAACAAUACCGCCUUAUAUAAUAUCAAA .........((.--..--.((((((.....-.(((((((......))))))---)...(((((((((((((....)))))))))))))((((..........)))).))))))..))... ( -28.90) >sp_ag.0 230687 116 + 2160267/0-120 CUCCUAUACGAA--UUCGUUUUUGUUUACAGGUGUGGAUGUUCCGGUCCAC--GAGAGUAUUUGAAAGGUUCCGGGGCCUUACAAAUGGGGUAAACAAUACCGCUUAUAAUCAUAUCAAA .......(((..--..)))..((((((((...(((((((......))))))--)....((((((.((((((....)))))).))))))..))))))))...................... ( -27.80) >sp_pn.0 272273 120 + 2038615/0-120 CUCCUAUAUAUAGUUCGAUUUUUGUUUACGGUGAUGGGUGUUCCUUCCCAUCGAAAAGUAUUUGGAAGGUUCCGGGGCCUUUCAAAUGGGGUAAACAAUACCGCCUACUAUCAUACCAAA ......((((((((.((....((((((((..(((((((........))))))).....(((((((((((((....)))))))))))))..))))))))...))...))))).)))..... ( -36.20) >consensus CUCCUAUACGAA__UC__UUUUUGUUUACAGAUGUGGAUGUUCCGGUCCAC___AAAGUAUUUGAAAGGUUCCGGGGCCUUUCAAAUGGGGUAAACAAUACCGCCUAAUAUCAUAUCAAA ......................(((((((....((((((......)))))).......(((((((((((((....)))))))))))))..)))))))....................... (-25.78 = -25.57 + -0.22) # Strand winner: reverse (1.00)

| Location | 169,680 – 169,792 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.49 |
| Mean single sequence MFE | -30.87 |
| Consensus MFE | -25.78 |
| Energy contribution | -25.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
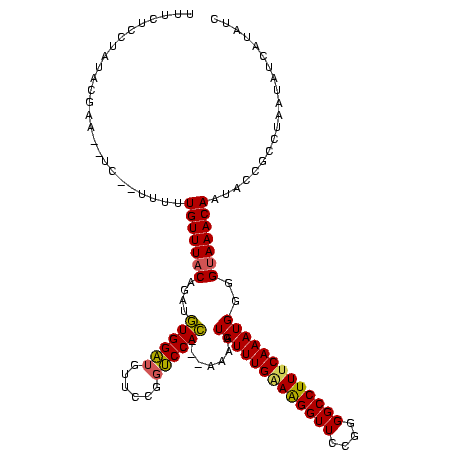
>sp_mu.0 169680 112 + 2030921/3-123 UUUCUCCUAUACGAA--UC--UUUAUGUUUACA-AUGUGGAUGUUCCGGUCCAC---AAAUUAUUUGAAGGGUUCCGGGGCCUUUCAAAUGGGGCAAACAAUACCGCCUUAUAUAAUAUC ...............--..--.((((((.....-.(((((((......))))))---)...(((((((((((((....)))))))))))))((((..........)))).)))))).... ( -28.50) >sp_ag.0 230687 116 + 2160267/3-123 UUUCUCCUAUACGAA--UUCGUUUUUGUUUACAGGUGUGGAUGUUCCGGUCCAC--GAGAGUAUUUGAAAGGUUCCGGGGCCUUACAAAUGGGGUAAACAAUACCGCUUAUAAUCAUAUC .(((((((..(((((--......)))))....))..((((((......))))))--))))).(((((.((((((....)))))).)))))(.((((.....)))).)............. ( -27.90) >sp_pn.0 272273 120 + 2038615/3-123 UUUCUCCUAUAUAUAGUUCGAUUUUUGUUUACGGUGAUGGGUGUUCCUUCCCAUCGAAAAGUAUUUGGAAGGUUCCGGGGCCUUUCAAAUGGGGUAAACAAUACCGCCUACUAUCAUACC .........((((((((.((....((((((((..(((((((........))))))).....(((((((((((((....)))))))))))))..))))))))...))...))))).))).. ( -36.20) >consensus UUUCUCCUAUACGAA__UC__UUUUUGUUUACAGAUGUGGAUGUUCCGGUCCAC___AAAGUAUUUGAAAGGUUCCGGGGCCUUUCAAAUGGGGUAAACAAUACCGCCUAAUAUCAUAUC .........................(((((((....((((((......)))))).......(((((((((((((....)))))))))))))..))))))).................... (-25.78 = -25.57 + -0.22) # Strand winner: reverse (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:50 2006