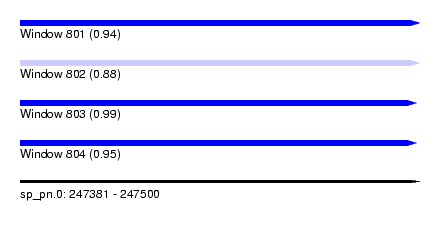
| Sequence ID | sp_pn.0 |
|---|---|
| Location | 247,381 – 247,500 |
| Length | 119 |
| Max. P | 0.989135 |

| Location | 247,381 – 247,500 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -25.85 |
| Energy contribution | -23.97 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 247381 119 + 2038615/40-160 AAUAAUAGACGGUACUUUUUACUUUUGGUCUCUCAAGAGUGUACAGGGACGUGCUGACAAAUGUUGCAAAAGUACACACAGAUGAUAUCUGUCACCAAGUGUAUCAUCACC-AAAAAUAA ...........(((((((((((((((((....)))))))))....(.(((((........))))).))))))))).....((((((((((.......)).))))))))...-........ ( -28.20) >sp_mu.0 333916 119 + 2030921/40-160 UAUGAUAAAUGGUACUUUUUACUUUUGGUCUCUCAAAAGUGUACAGAGACGUGCUGACAAAUGUUGCAAA-GUACACACAGAUUAGGACUGUCACCAAGUGCCUAAUCACCCAAAAAUAA .........(((((((((.(((((((((....)))))))))....(..((((........))))..))))-)))).))..((((((((((.......))).)))))))............ ( -27.80) >sp_ag.0 393162 119 - 2160267/40-160 UAUGAUAAACGGUACUUUU-ACUUUUGGUCUCUCAAAAGUGUACAGAGACGUGCUGACAAUUGUUGCAAAAGUACACGCAGAUAUAGGCUGUCACCAAGUGCUAUAUCAACCAAAAAUAA .........((((((((((-.......((((((...........))))))..((.(((....))))))))))))).))..((((((((((.......))).)))))))............ ( -27.00) >consensus UAUGAUAAACGGUACUUUUUACUUUUGGUCUCUCAAAAGUGUACAGAGACGUGCUGACAAAUGUUGCAAAAGUACACACAGAUAAAGACUGUCACCAAGUGCAUAAUCACCCAAAAAUAA ...........((((((((.((((((((....))))))))((((......)))).............)))))))).....((((((((((.......))).)))))))............ (-25.85 = -23.97 + -1.88)

| Location | 247,381 – 247,500 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.94 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -26.49 |
| Energy contribution | -24.93 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876131 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 247381 119 + 2038615/40-160 UUAUUUUU-GGUGAUGAUACACUUGGUGACAGAUAUCAUCUGUGUGUACUUUUGCAACAUUUGUCAGCACGUCCCUGUACACUCUUGAGAGACCAAAAGUAAAAAGUACCGUCUAUUAUU ((((((((-((((((((((...(((....))).))))))).(.((((((...(((.((....))..))).......)))))).).......))))))))))).................. ( -28.80) >sp_mu.0 333916 119 + 2030921/40-160 UUAUUUUUGGGUGAUUAGGCACUUGGUGACAGUCCUAAUCUGUGUGUAC-UUUGCAACAUUUGUCAGCACGUCUCUGUACACUUUUGAGAGACCAAAAGUAAAAAGUACCAUUUAUCAUA ((((((((((..(((((((.(((.(....)))))))))))...((((((-..(((.((....))..))).......))))))..........)))))))))).................. ( -30.40) >sp_ag.0 393162 119 - 2160267/40-160 UUAUUUUUGGUUGAUAUAGCACUUGGUGACAGCCUAUAUCUGCGUGUACUUUUGCAACAAUUGUCAGCACGUCUCUGUACACUUUUGAGAGACCAAAAGU-AAAAGUACCGUUUAUCAUA .......((((.(((((((...(((....))).))))))).(((.((((((((((......((....)).((((((...........)))))).....))-)))))))))))..)))).. ( -30.90) >consensus UUAUUUUUGGGUGAUAAAGCACUUGGUGACAGACAUAAUCUGUGUGUACUUUUGCAACAUUUGUCAGCACGUCUCUGUACACUUUUGAGAGACCAAAAGUAAAAAGUACCGUUUAUCAUA ((((((((((..(((((((...(((....))).)))))))...((((((...(((.((....))..))).......))))))..........)))))))))).................. (-26.49 = -24.93 + -1.55) # Strand winner: reverse (0.76)

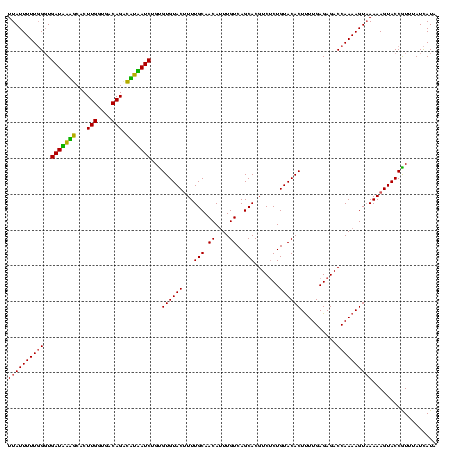
| Location | 247,381 – 247,499 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -26.83 |
| Energy contribution | -24.83 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 247381 118 + 2038615/56-176 UUUUACUUUUGGUCUCUCAAGAGUGUACAGGGACGUGCUGACAAAUGUUGCAAAAGUACACACAGAUGAUAUCUGUCACCAAGUGUAUCAUCACC-AAAAAUAAAAAAAC-ACAGGAGAA (((((.(((((((.........((((((.(.(((((........))))).)....))))))...((((((((((.......)).)))))))))))-)))).)))))....-......... ( -28.90) >sp_mu.0 333916 118 + 2030921/56-176 UUUUACUUUUGGUCUCUCAAAAGUGUACAGAGACGUGCUGACAAAUGUUGCAAA-GUACACACAGAUUAGGACUGUCACCAAGUGCCUAAUCACCCAAAAAUAAAAAAAU-ACAGGAGAA (((((.((((((..........((((((...(((((........))))).....-))))))...((((((((((.......))).)))))))..)))))).)))))....-......... ( -29.50) >sp_ag.0 393162 119 - 2160267/56-176 UUU-ACUUUUGGUCUCUCAAAAGUGUACAGAGACGUGCUGACAAUUGUUGCAAAAGUACACGCAGAUAUAGGCUGUCACCAAGUGCUAUAUCAACCAAAAAUAAAAAACUUACAGGAGAA (((-(.(((((((.........((((((.......(((.(((....))))))...))))))...((((((((((.......))).))))))).))))))).))))............... ( -26.90) >consensus UUUUACUUUUGGUCUCUCAAAAGUGUACAGAGACGUGCUGACAAAUGUUGCAAAAGUACACACAGAUAAAGACUGUCACCAAGUGCAUAAUCACCCAAAAAUAAAAAAAU_ACAGGAGAA .....((((((...........((((((.......(((.(((....))))))...))))))...((((((((((.......))).)))))))....................)))))).. (-26.83 = -24.83 + -2.00)

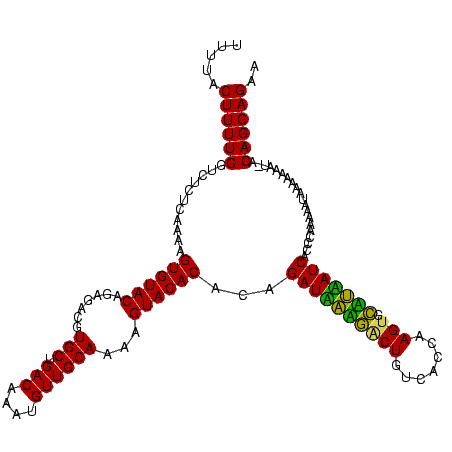
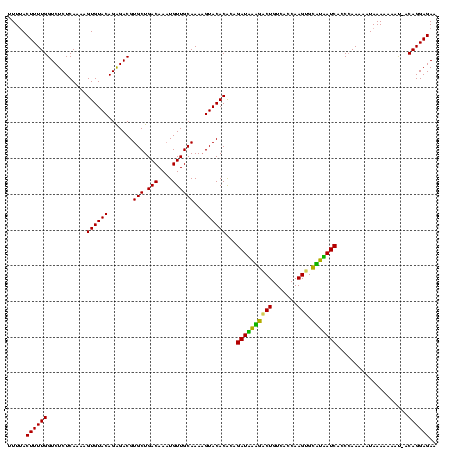
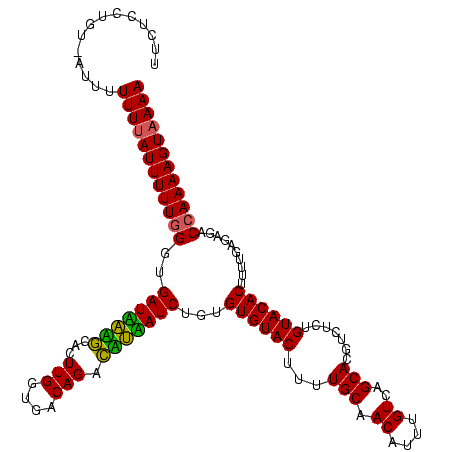
| Location | 247,381 – 247,499 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -27.69 |
| Energy contribution | -26.13 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 247381 118 + 2038615/56-176 UUCUCCUGU-GUUUUUUUAUUUUU-GGUGAUGAUACACUUGGUGACAGAUAUCAUCUGUGUGUACUUUUGCAACAUUUGUCAGCACGUCCCUGUACACUCUUGAGAGACCAAAAGUAAAA .........-....((((((((((-((((((((((...(((....))).))))))).(.((((((...(((.((....))..))).......)))))).).......))))))))))))) ( -30.00) >sp_mu.0 333916 118 + 2030921/56-176 UUCUCCUGU-AUUUUUUUAUUUUUGGGUGAUUAGGCACUUGGUGACAGUCCUAAUCUGUGUGUAC-UUUGCAACAUUUGUCAGCACGUCUCUGUACACUUUUGAGAGACCAAAAGUAAAA .........-....((((((((((((..(((((((.(((.(....)))))))))))...((((((-..(((.((....))..))).......))))))..........)))))))))))) ( -31.60) >sp_ag.0 393162 119 - 2160267/56-176 UUCUCCUGUAAGUUUUUUAUUUUUGGUUGAUAUAGCACUUGGUGACAGCCUAUAUCUGCGUGUACUUUUGCAACAAUUGUCAGCACGUCUCUGUACACUUUUGAGAGACCAAAAGU-AAA ...............((((((((((((((((((((...(((....))).)))))))...((((((...(((.((....))..))).......))))))........))))))))))-))) ( -29.90) >consensus UUCUCCUGU_AUUUUUUUAUUUUUGGGUGAUAAAGCACUUGGUGACAGACAUAAUCUGUGUGUACUUUUGCAACAUUUGUCAGCACGUCUCUGUACACUUUUGAGAGACCAAAAGUAAAA ..............((((((((((((..(((((((...(((....))).)))))))...((((((...(((.((....))..))).......))))))..........)))))))))))) (-27.69 = -26.13 + -1.55) # Strand winner: reverse (0.88)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:49 2006