| Sequence ID | sp_mu.0 |
|---|---|
| Location | 145,538 – 1,146,731 |
| Length | 1001193 |
| Max. P | 0.999778 |

| Location | 1,146,657 – 1,146,731 |
|---|---|
| Length | 74 |
| Sequences | 3 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -15.70 |
| Consensus MFE | -15.07 |
| Energy contribution | -13.30 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
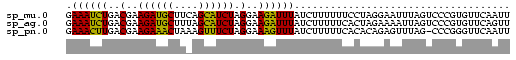
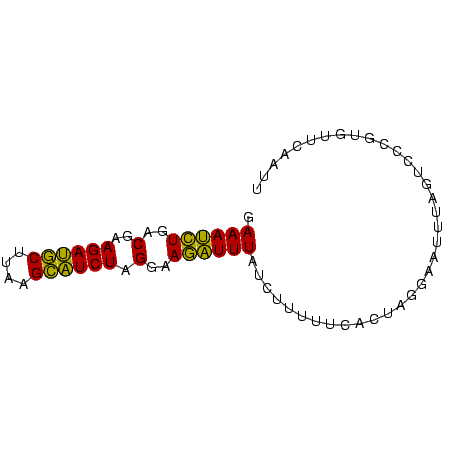
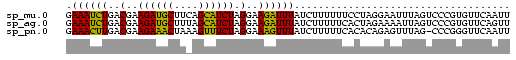
>sp_mu.0 1146657 74 + 2030921 GAAAUCUGACGAAGAUGCUUCAGCAUCUAGGAAGAUUUAUCUUUUUUCCUAGGAAUUUAGUCCCGUGUUCAAUU (((....(((...(((((....)))))((((((((........))))))))........))).....))).... ( -18.70) >sp_ag.0 2100870 74 - 2160267 GAAAUCUGACGAAGAUGCUUUAGCAUCUAGGAAGAUUUAUCUUUUUCACUAGAAAAUUAGUCCCGUGUUCAGUU .((((((..(..((((((....)))))).)..))))))....(((((....))))).................. ( -15.60) >sp_pn.0 202492 73 + 2038615 GAAACUUGACGAAGAAACUAAAGUUUCUAGGAAAGUUUAUCUUUUUCACACAGAGUUUAG-CCCGGGUUCAAUU .((((((..(..((((((....)))))).)..))))))...............((((.((-(....))).)))) ( -12.80) >consensus GAAAUCUGACGAAGAUGCUUAAGCAUCUAGGAAGAUUUAUCUUUUUCACUAGGAAUUUAGUCCCGUGUUCAAUU .((((((..(..((((((....)))))).)..)))))).................................... (-15.07 = -13.30 + -1.77)

| Location | 1,146,657 – 1,146,731 |
|---|---|
| Length | 74 |
| Sequences | 3 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -13.57 |
| Consensus MFE | -11.88 |
| Energy contribution | -10.00 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
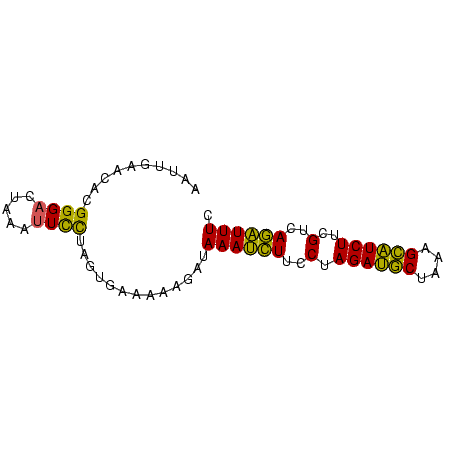
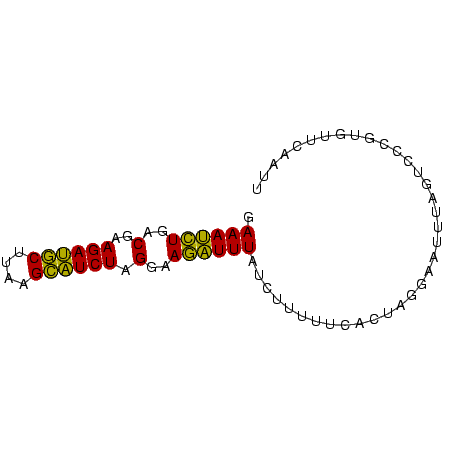
>sp_mu.0 1146657 74 + 2030921 AAUUGAACACGGGACUAAAUUCCUAGGAAAAAAGAUAAAUCUUCCUAGAUGCUGAAGCAUCUUCGUCAGAUUUC ..((((....((((......))))(((((............)))))((((((....))))))...))))..... ( -14.20) >sp_ag.0 2100870 74 - 2160267 AACUGAACACGGGACUAAUUUUCUAGUGAAAAAGAUAAAUCUUCCUAGAUGCUAAAGCAUCUUCGUCAGAUUUC ..((((....((((....(((((....)))))(((....)))))))((((((....))))))...))))..... ( -16.20) >sp_pn.0 202492 73 + 2038615 AAUUGAACCCGGG-CUAAACUCUGUGUGAAAAAGAUAAACUUUCCUAGAAACUUUAGUUUCUUCGUCAAGUUUC ..((..((.((((-......)))).))..)).....((((((..(.((((((....))))))..)..)))))). ( -10.30) >consensus AAUUGAACACGGGACUAAAUUCCUAGUGAAAAAGAUAAAUCUUCCUAGAUGCUAAAGCAUCUUCGUCAGAUUUC ..........((((.....)))).............((((((..(.((((((....))))))..)..)))))). (-11.88 = -10.00 + -1.88) # Strand winner: forward (0.99)


| Location | 1,146,657 – 1,146,731 |
|---|---|
| Length | 74 |
| Sequences | 3 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -15.70 |
| Consensus MFE | -15.07 |
| Energy contribution | -13.30 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1146657 74 + 2030921/0-74 GAAAUCUGACGAAGAUGCUUCAGCAUCUAGGAAGAUUUAUCUUUUUUCCUAGGAAUUUAGUCCCGUGUUCAAUU (((....(((...(((((....)))))((((((((........))))))))........))).....))).... ( -18.70) >sp_ag.0 2100870 74 - 2160267/0-74 GAAAUCUGACGAAGAUGCUUUAGCAUCUAGGAAGAUUUAUCUUUUUCACUAGAAAAUUAGUCCCGUGUUCAGUU .((((((..(..((((((....)))))).)..))))))....(((((....))))).................. ( -15.60) >sp_pn.0 202492 73 + 2038615/0-74 GAAACUUGACGAAGAAACUAAAGUUUCUAGGAAAGUUUAUCUUUUUCACACAGAGUUUAG-CCCGGGUUCAAUU .((((((..(..((((((....)))))).)..))))))...............((((.((-(....))).)))) ( -12.80) >consensus GAAAUCUGACGAAGAUGCUUAAGCAUCUAGGAAGAUUUAUCUUUUUCACUAGGAAUUUAGUCCCGUGUUCAAUU .((((((..(..((((((....)))))).)..)))))).................................... (-15.07 = -13.30 + -1.77)

| Location | 1,146,657 – 1,146,731 |
|---|---|
| Length | 74 |
| Sequences | 3 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -13.57 |
| Consensus MFE | -11.88 |
| Energy contribution | -10.00 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1146657 74 + 2030921/0-74 AAUUGAACACGGGACUAAAUUCCUAGGAAAAAAGAUAAAUCUUCCUAGAUGCUGAAGCAUCUUCGUCAGAUUUC ..((((....((((......))))(((((............)))))((((((....))))))...))))..... ( -14.20) >sp_ag.0 2100870 74 - 2160267/0-74 AACUGAACACGGGACUAAUUUUCUAGUGAAAAAGAUAAAUCUUCCUAGAUGCUAAAGCAUCUUCGUCAGAUUUC ..((((....((((....(((((....)))))(((....)))))))((((((....))))))...))))..... ( -16.20) >sp_pn.0 202492 73 + 2038615/0-74 AAUUGAACCCGGG-CUAAACUCUGUGUGAAAAAGAUAAACUUUCCUAGAAACUUUAGUUUCUUCGUCAAGUUUC ..((..((.((((-......)))).))..)).....((((((..(.((((((....))))))..)..)))))). ( -10.30) >consensus AAUUGAACACGGGACUAAAUUCCUAGUGAAAAAGAUAAAUCUUCCUAGAUGCUAAAGCAUCUUCGUCAGAUUUC ..........((((.....)))).............((((((..(.((((((....))))))..)..)))))). (-11.88 = -10.00 + -1.88) # Strand winner: forward (0.99)

| Location | 145,538 – 145,612 |
|---|---|
| Length | 74 |
| Sequences | 3 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -23.93 |
| Energy contribution | -20.83 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.53 |
| Mean z-score | -3.11 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
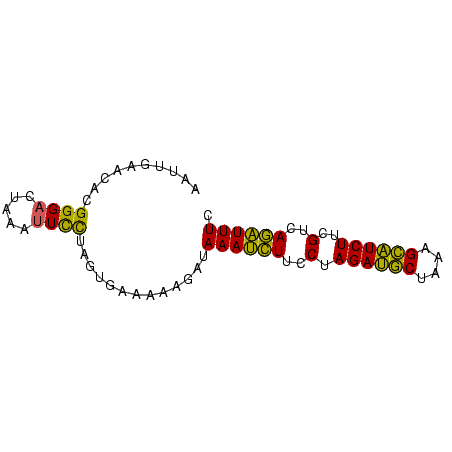
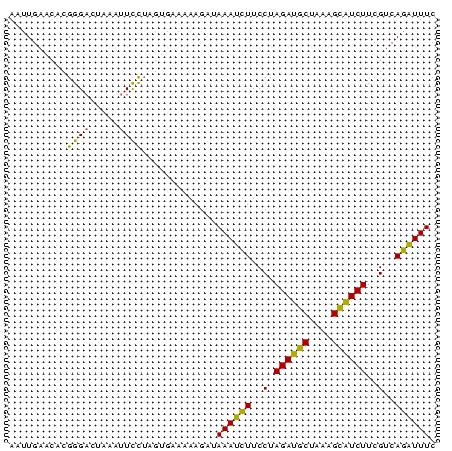
>sp_mu.0 145538 74 - 2030921 UUAGUAUAGGCGAGUUUUUAUAGGGAACA-CCUUUUCCCCUAUAAAGGCGCUAGCAUUUUACAAAUAAGGAGAAA ...(((...((..((((((((((((....-.......))))))))))))....))....)))............. ( -19.10) >sp_ag.0 85398 74 + 2160267 AUCGUAUAGGCGAGUCUUUGCGGGGAGUA-CCUUUUCCCUCGUAAAGGCGUUAGCAUUUUACAAAUAAGGAGAAA ...(((...((..((((((((((((((..-....))))).)))))))))....))....)))............. ( -25.10) >sp_pn.0 205074 75 + 2038615 AUCGUAUAGGCGAGUUGAUGGGGGAGACAACCUUUUCUCCCUUAUCGGCGCUAGCAUUUUACAAAAGAGGAGAAA ...(((...((..((((((((((((((........))))))))))))))....))....)))............. ( -26.10) >consensus AUCGUAUAGGCGAGUUUUUGCGGGGAACA_CCUUUUCCCCCAUAAAGGCGCUAGCAUUUUACAAAUAAGGAGAAA ...(((...((..((((((((((((............))))))))))))....))....)))............. (-23.93 = -20.83 + -3.10)

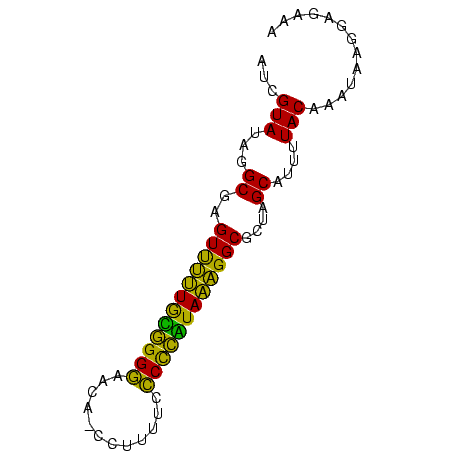
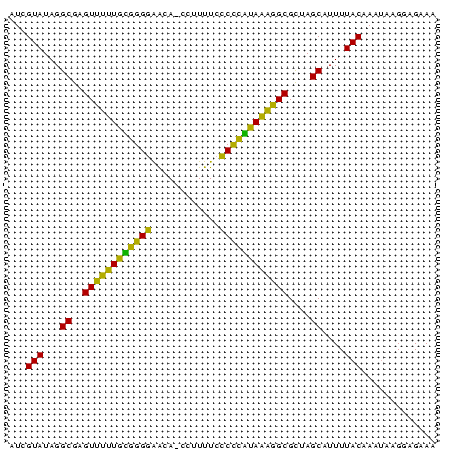
| Location | 145,538 – 145,612 |
|---|---|
| Length | 74 |
| Sequences | 3 |
| Columns | 75 |
| Reading direction | forward |
| Mean pairwise identity | 79.46 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -23.93 |
| Energy contribution | -20.83 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.53 |
| Mean z-score | -3.11 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 145538 74 - 2030921/0-75 UUAGUAUAGGCGAGUUUUUAUAGGGAACA-CCUUUUCCCCUAUAAAGGCGCUAGCAUUUUACAAAUAAGGAGAAA ...(((...((..((((((((((((....-.......))))))))))))....))....)))............. ( -19.10) >sp_ag.0 85398 74 + 2160267/0-75 AUCGUAUAGGCGAGUCUUUGCGGGGAGUA-CCUUUUCCCUCGUAAAGGCGUUAGCAUUUUACAAAUAAGGAGAAA ...(((...((..((((((((((((((..-....))))).)))))))))....))....)))............. ( -25.10) >sp_pn.0 205074 75 + 2038615/0-75 AUCGUAUAGGCGAGUUGAUGGGGGAGACAACCUUUUCUCCCUUAUCGGCGCUAGCAUUUUACAAAAGAGGAGAAA ...(((...((..((((((((((((((........))))))))))))))....))....)))............. ( -26.10) >consensus AUCGUAUAGGCGAGUUUUUGCGGGGAACA_CCUUUUCCCCCAUAAAGGCGCUAGCAUUUUACAAAUAAGGAGAAA ...(((...((..((((((((((((............))))))))))))....))....)))............. (-23.93 = -20.83 + -3.10)

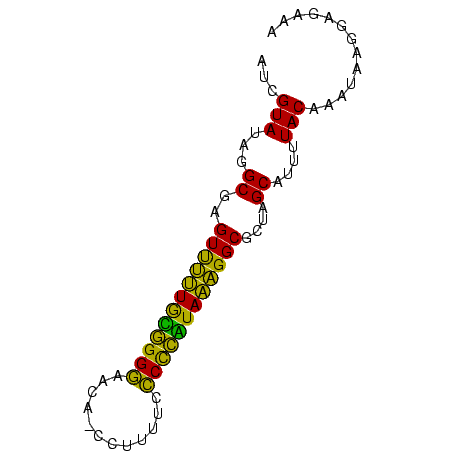
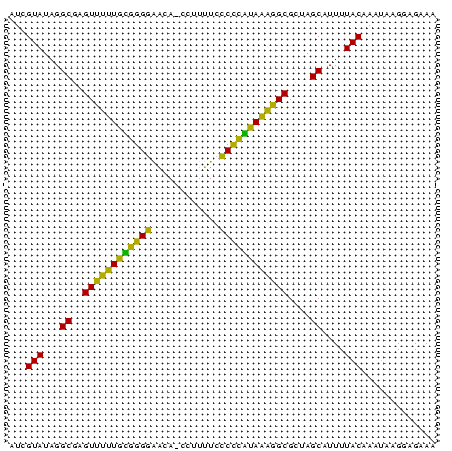
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:45 2006