| Sequence ID | sp_mu.0 |
|---|---|
| Location | 138,040 – 138,106 |
| Length | 66 |
| Max. P | 0.999396 |

| Location | 138,040 – 138,106 |
|---|---|
| Length | 66 |
| Sequences | 3 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -10.10 |
| Consensus MFE | -6.59 |
| Energy contribution | -7.27 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940424 |
| Prediction | RNA |
| WARNING | Sequence 3: Base composition out of range. |
Download alignment: ClustalW | MAF
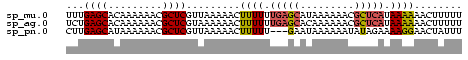
>sp_mu.0 138040 66 - 2030921 UUUGAGCACAAAAAACGCUCGUUAAAAACUUUUUUGAGCAUAAAAAACGCUCAUAAAAAACUUUUU ...((((.........)))).........((((.(((((.........))))).))))........ ( -11.40) >sp_ag.0 76603 66 + 2160267 UCUGAGCACAAAAAACGCUCGUAAAAAACUUUUUUGAGCACAAAAAACGCUCAUAAAAAACUUUUU ...((((.........)))).........((((.(((((.........))))).))))........ ( -11.40) >sp_pn.0 196008 63 + 2038615 CUUGAGCAUAAAAAACGCUCGUUAAAAACUUUUU---GAAUAAAAAAUAUAGAAAAGGAACUAUUU ...((((.........))))(((.....((((((---..(((.....))).)))))).)))..... ( -7.50) >consensus UUUGAGCACAAAAAACGCUCGUUAAAAACUUUUUUGAGCAUAAAAAACGCUCAUAAAAAACUUUUU ...((((.........)))).........((((.(((((.........))))).))))........ ( -6.59 = -7.27 + 0.67)
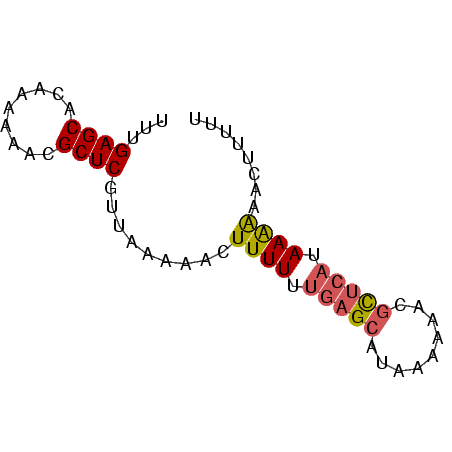
| Location | 138,040 – 138,106 |
|---|---|
| Length | 66 |
| Sequences | 3 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -14.33 |
| Consensus MFE | -9.59 |
| Energy contribution | -11.60 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969298 |
| Prediction | RNA |
| WARNING | Sequence 3: Base composition out of range. |
Download alignment: ClustalW | MAF
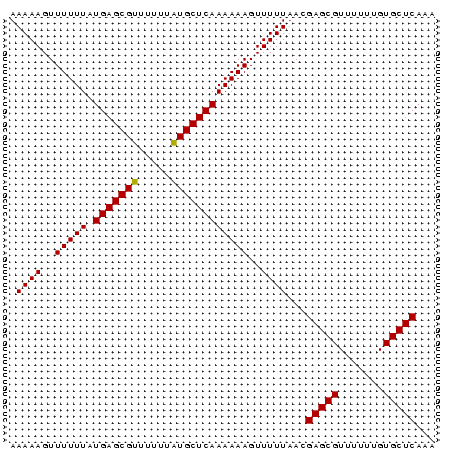
>sp_mu.0 138040 66 - 2030921 AAAAAGUUUUUUAUGAGCGUUUUUUAUGCUCAAAAAAGUUUUUAACGAGCGUUUUUUGUGCUCAAA .((((..(((((.(((((((.....))))))))))))..))))...(((((.......)))))... ( -17.80) >sp_ag.0 76603 66 + 2160267 AAAAAGUUUUUUAUGAGCGUUUUUUGUGCUCAAAAAAGUUUUUUACGAGCGUUUUUUGUGCUCAGA (((((..(((((.((((((.......)))))))))))..)))))..(((((.......)))))... ( -16.30) >sp_pn.0 196008 63 + 2038615 AAAUAGUUCCUUUUCUAUAUUUUUUAUUC---AAAAAGUUUUUAACGAGCGUUUUUUAUGCUCAAG ..((((........))))(((((((....---))))))).......((((((.....))))))... ( -8.90) >consensus AAAAAGUUUUUUAUGAGCGUUUUUUAUGCUCAAAAAAGUUUUUAACGAGCGUUUUUUGUGCUCAAA (((((.(((((..(((((((.....)))))))))))).)))))...((((((.....))))))... ( -9.59 = -11.60 + 2.01) # Strand winner: reverse (0.98)
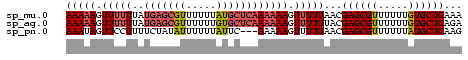
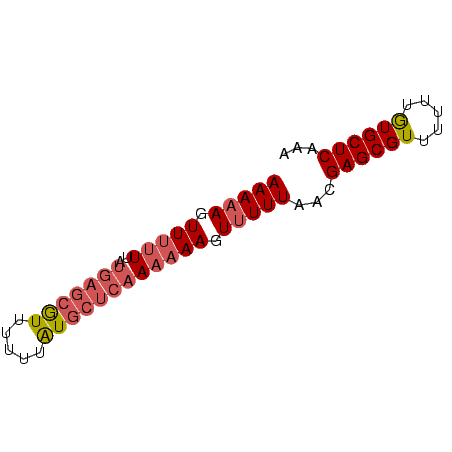
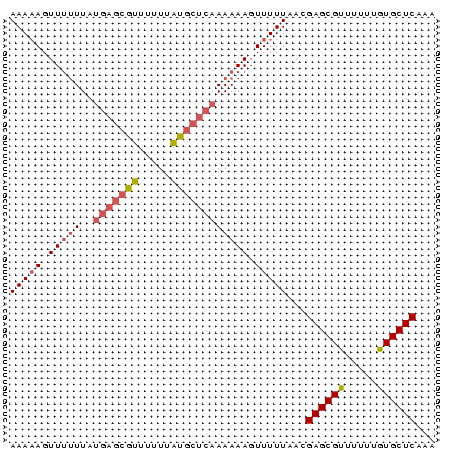
| Location | 138,040 – 138,106 |
|---|---|
| Length | 66 |
| Sequences | 2 |
| Columns | 66 |
| Reading direction | forward |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -11.40 |
| Consensus MFE | -11.40 |
| Energy contribution | -11.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.35 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 138040 66 - 2030921/0-66 UUUGAGCACAAAAAACGCUCGUUAAAAACUUUUUUGAGCAUAAAAAACGCUCAUAAAAAACUUUUU ...((((.........)))).........((((.(((((.........))))).))))........ ( -11.40) >sp_ag.0 76603 66 + 2160267/0-66 UCUGAGCACAAAAAACGCUCGUAAAAAACUUUUUUGAGCACAAAAAACGCUCAUAAAAAACUUUUU ...((((.........)))).........((((.(((((.........))))).))))........ ( -11.40) >consensus UCUGAGCACAAAAAACGCUCGUAAAAAACUUUUUUGAGCACAAAAAACGCUCAUAAAAAACUUUUU ...((((.........)))).........((((.(((((.........))))).))))........ (-11.40 = -11.40 + 0.00)

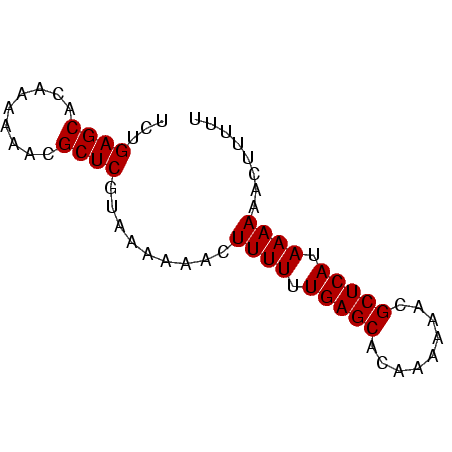
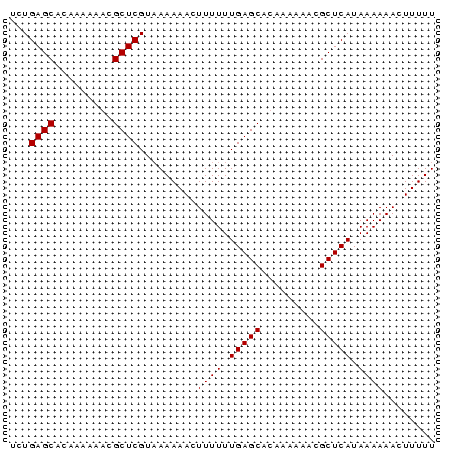
| Location | 138,040 – 138,106 |
|---|---|
| Length | 66 |
| Sequences | 2 |
| Columns | 66 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -17.05 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.15 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.45 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.19 |
| SVM RNA-class probability | 0.998700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 138040 66 - 2030921/0-66 AAAAAGUUUUUUAUGAGCGUUUUUUAUGCUCAAAAAAGUUUUUAACGAGCGUUUUUUGUGCUCAAA .((((..(((((.(((((((.....))))))))))))..))))...(((((.......)))))... ( -17.80) >sp_ag.0 76603 66 + 2160267/0-66 AAAAAGUUUUUUAUGAGCGUUUUUUGUGCUCAAAAAAGUUUUUUACGAGCGUUUUUUGUGCUCAGA (((((..(((((.((((((.......)))))))))))..)))))..(((((.......)))))... ( -16.30) >consensus AAAAAGUUUUUUAUGAGCGUUUUUUAUGCUCAAAAAAGUUUUUAACGAGCGUUUUUUGUGCUCAAA .((((..(((((.(((((((.....))))))))))))..))))...(((((.......)))))... (-16.40 = -16.15 + -0.25) # Strand winner: reverse (0.99)

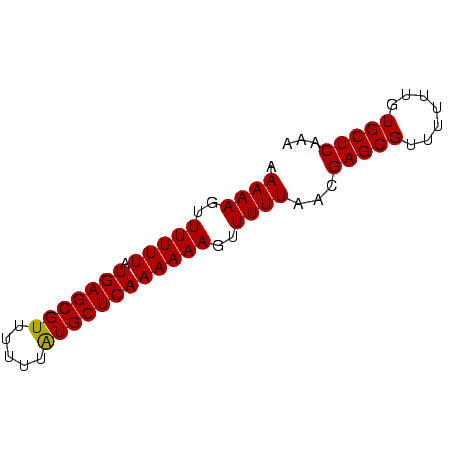
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:41 2006