
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 24,586 – 24,682 |
| Length | 96 |
| Max. P | 0.997853 |

| Location | 24,586 – 24,682 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.73 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.31 |
| Mean z-score | -5.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 24586 96 - 2030921 AAUUUGAUAAAAUAAAUAUAACAAUAAUCAUGAUGGUCAAAGCUCAUGAGAAUUGUUGGUCUUUU-GACCCACAAUUUUCGAGGGUUUUGGCUAUUU ...............................((((((((((((((.((((((((((.((((....-)))).)))))))))).)))))))))))))). ( -34.90) >sp_ag.0 34359 97 - 2160267 UAAAUGCUAAAAUAAAUUUAACAAUAACUAUGAUGGUCAAAGCUCAUGGAAUUUGUAAGUCUUUUUGACGUACAAUUUUCGAGAGUUUUGGCUAUUU ...............................((((((((((((((.(((((.(((((.(((.....))).))))).))))).)))))))))))))). ( -30.00) >sp_pn.0 131753 97 + 2038615 AAUUUGCUACAAUAAUAACAGCAAUAGAAAUGAUGGUCAAAGCUCAUGGAUGUUGCAGGCUUUUUUGUCCUGCACUUCUUUGUAGUUUUGACUGUUU ...(((((...........))))).......(((((((((((((((.(((.((.((((((......).))))))).))).)).))))))))))))). ( -30.50) >consensus AAUUUGCUAAAAUAAAUAUAACAAUAAAAAUGAUGGUCAAAGCUCAUGGAAAUUGUAGGUCUUUUUGACCUACAAUUUUCGAGAGUUUUGGCUAUUU ...............................((((((((((((((.(((((((((((((((.....))))))))))))))).)))))))))))))). (-29.72 = -30.73 + 1.02)


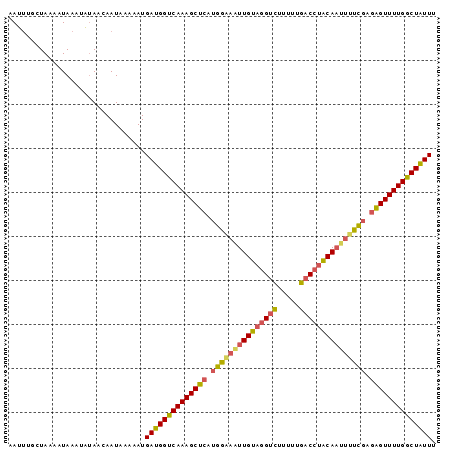
| Location | 24,586 – 24,682 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -9.82 |
| Energy contribution | -12.16 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 24586 96 - 2030921 AAAUAGCCAAAACCCUCGAAAAUUGUGGGUC-AAAAGACCAACAAUUCUCAUGAGCUUUGACCAUCAUGAUUAUUGUUAUAUUUAUUUUAUCAAAUU ..(((((.............((((((.((((-....)))).)))))).((((((..........)))))).....)))))................. ( -16.90) >sp_ag.0 34359 97 - 2160267 AAAUAGCCAAAACUCUCGAAAAUUGUACGUCAAAAAGACUUACAAAUUCCAUGAGCUUUGACCAUCAUAGUUAUUGUUAAAUUUAUUUUAGCAUUUA .((((((((((.(((..(((..(((((.(((.....))).))))).)))...))).)))).........))))))((((((.....))))))..... ( -16.50) >sp_pn.0 131753 97 + 2038615 AAACAGUCAAAACUACAAAGAAGUGCAGGACAAAAAAGCCUGCAACAUCCAUGAGCUUUGACCAUCAUUUCUAUUGCUGUUAUUAUUGUAGCAAAUU .....((((((.((.((..((..((((((.(......)))))))...))..)))).))))))...........((((((.........))))))... ( -21.90) >consensus AAAUAGCCAAAACUCUCGAAAAUUGUAGGUCAAAAAGACCUACAAAUUCCAUGAGCUUUGACCAUCAUAAUUAUUGUUAUAUUUAUUUUAGCAAAUU .......((((.(((........((((((((.....))))))))........))).)))).............((((((((.....))))))))... ( -9.82 = -12.16 + 2.34) # Strand winner: forward (0.98)

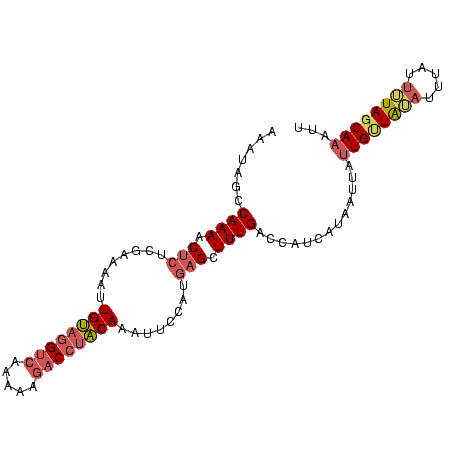
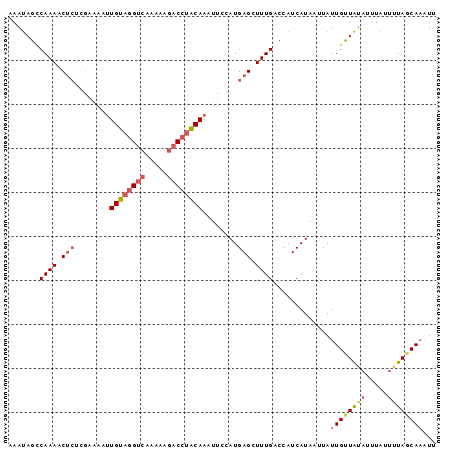
| Location | 24,586 – 24,682 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -29.72 |
| Energy contribution | -30.73 |
| Covariance contribution | 1.02 |
| Combinations/Pair | 1.31 |
| Mean z-score | -5.33 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 24586 96 - 2030921/0-97 AAUUUGAUAAAAUAAAUAUAACAAUAAUCAUGAUGGUCAAAGCUCAUGAGAAUUGUUGGUCUUUU-GACCCACAAUUUUCGAGGGUUUUGGCUAUUU ...............................((((((((((((((.((((((((((.((((....-)))).)))))))))).)))))))))))))). ( -34.90) >sp_ag.0 34359 97 - 2160267/0-97 UAAAUGCUAAAAUAAAUUUAACAAUAACUAUGAUGGUCAAAGCUCAUGGAAUUUGUAAGUCUUUUUGACGUACAAUUUUCGAGAGUUUUGGCUAUUU ...............................((((((((((((((.(((((.(((((.(((.....))).))))).))))).)))))))))))))). ( -30.00) >sp_pn.0 131753 97 + 2038615/0-97 AAUUUGCUACAAUAAUAACAGCAAUAGAAAUGAUGGUCAAAGCUCAUGGAUGUUGCAGGCUUUUUUGUCCUGCACUUCUUUGUAGUUUUGACUGUUU ...(((((...........))))).......(((((((((((((((.(((.((.((((((......).))))))).))).)).))))))))))))). ( -30.50) >consensus AAUUUGCUAAAAUAAAUAUAACAAUAAAAAUGAUGGUCAAAGCUCAUGGAAAUUGUAGGUCUUUUUGACCUACAAUUUUCGAGAGUUUUGGCUAUUU ...............................((((((((((((((.(((((((((((((((.....))))))))))))))).)))))))))))))). (-29.72 = -30.73 + 1.02)

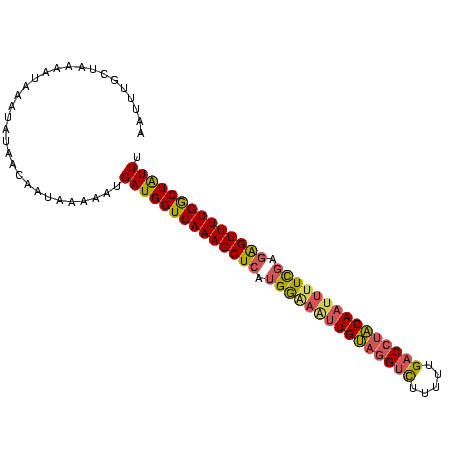

| Location | 24,586 – 24,682 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.23 |
| Mean single sequence MFE | -18.43 |
| Consensus MFE | -9.82 |
| Energy contribution | -12.16 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 24586 96 - 2030921/0-97 AAAUAGCCAAAACCCUCGAAAAUUGUGGGUC-AAAAGACCAACAAUUCUCAUGAGCUUUGACCAUCAUGAUUAUUGUUAUAUUUAUUUUAUCAAAUU ..(((((.............((((((.((((-....)))).)))))).((((((..........)))))).....)))))................. ( -16.90) >sp_ag.0 34359 97 - 2160267/0-97 AAAUAGCCAAAACUCUCGAAAAUUGUACGUCAAAAAGACUUACAAAUUCCAUGAGCUUUGACCAUCAUAGUUAUUGUUAAAUUUAUUUUAGCAUUUA .((((((((((.(((..(((..(((((.(((.....))).))))).)))...))).)))).........))))))((((((.....))))))..... ( -16.50) >sp_pn.0 131753 97 + 2038615/0-97 AAACAGUCAAAACUACAAAGAAGUGCAGGACAAAAAAGCCUGCAACAUCCAUGAGCUUUGACCAUCAUUUCUAUUGCUGUUAUUAUUGUAGCAAAUU .....((((((.((.((..((..((((((.(......)))))))...))..)))).))))))...........((((((.........))))))... ( -21.90) >consensus AAAUAGCCAAAACUCUCGAAAAUUGUAGGUCAAAAAGACCUACAAAUUCCAUGAGCUUUGACCAUCAUAAUUAUUGUUAUAUUUAUUUUAGCAAAUU .......((((.(((........((((((((.....))))))))........))).)))).............((((((((.....))))))))... ( -9.82 = -12.16 + 2.34) # Strand winner: forward (0.98)

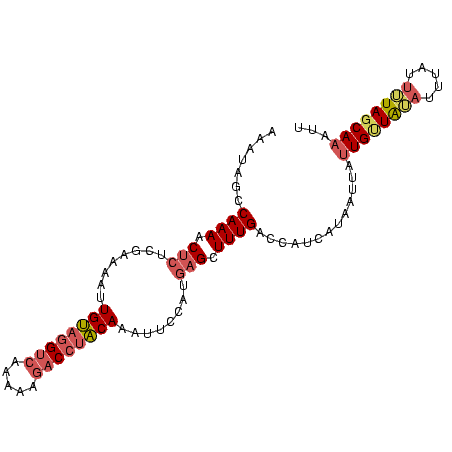
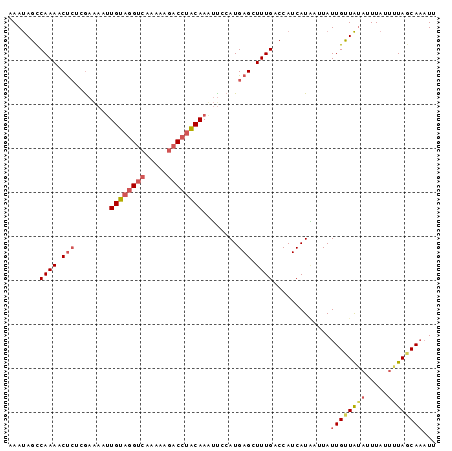
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:38 2006