
| Sequence ID | sy_au.0 |
|---|---|
| Location | 577,882 – 578,002 |
| Length | 120 |
| Max. P | 0.999906 |

| Location | 577,882 – 577,979 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.26 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -6.99 |
| Energy contribution | -9.77 |
| Covariance contribution | 2.78 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.770703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 97 + 2809422/40-160 CCAAAA--AACGU--AACUAUAAGUU----ACAAACAUUAU------------UUAGUAUUU-AUGAGCUAAUCAAACAUCAUAAUU--UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUG ((....--...((--(((.....)))----)).........------------.........-.((((((((((((((.(((((...--..)))))...)))))))..)))))))))... ( -23.20) >sy_sa.0 210243 120 - 2516575/40-160 CCGAACGCAACAUUAAAUUGUUGGUUCAAAACAAGUGUUAGAGAUAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUG ((((((.(((((......))))))))).......((((((....))))))..............((((((((((((((.((((((.....))))))...)))))))..)))))))))... ( -35.90) >sp_ag.0 91093 116 + 2160267/40-160 UGCAGGGUGGAUUUACAUUGUAAAUCCGUCAAUGACAAAAAACAAUAAAUCUGUCAGCGAGACA-GAAAAGAGCAAAGCUCAAACUU---UUAAUGAGAGUUUGAUCCUGGCUCAGGACG ........((((((((...))))))))(((...................((((((.....))))-))...((((...(.((((((((---((...)))))))))).)...))))..))). ( -34.90) >consensus CCAAAAG_AACAUUAAAUUGUAAGUUC___ACAAACAUUAAA_A__A_A____UUAGUAUUU_AUGAGCUAAUCAAACAUCAUAAUU__UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUG .................................................................(((((((((((((.((((((.....))))))...)))))))..))))))...... ( -6.99 = -9.77 + 2.78)


| Location | 577,882 – 577,979 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 57.26 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -8.46 |
| Energy contribution | -9.02 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.30 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 97 + 2809422/40-160 CAUCCUGAGCCAGGAUCAAACUCUCCAUAAA--AAUUAUGAUGUUUGAUUAGCUCAU-AAAUACUAA------------AUAAUGUUUGU----AACUUAUAGUU--ACGUU--UUUUGG .....(((((...((((((((....((((..--...))))..)))))))).))))).-.....((((------------(.......(((----(((.....)))--)))..--.))))) ( -20.70) >sy_sa.0 210243 120 - 2516575/40-160 CAUCCUGAGCCAGGAUCAAACUCUCCAUAAAUGAAUUAUGAUGUUUGAUUAGCUCAUAAAAUACUAAUUUGUGUUAUCUCUAACACUUGUUUUGAACCAACAAUUUAAUGUUGCGUUCGG .....(((((...((((((((....(((((.....)))))..)))))))).))))).........(((..((((((....))))))..))).((((((((((......))))).))))). ( -30.90) >sp_ag.0 91093 116 + 2160267/40-160 CGUCCUGAGCCAGGAUCAAACUCUCAUUAA---AAGUUUGAGCUUUGCUCUUUUC-UGUCUCGCUGACAGAUUUAUUGUUUUUUGUCAUUGACGGAUUUACAAUGUAAAUCCACCCUGCA .(((..((((.(((.(((((((........---.))))))).))).))))...((-((((.....))))))...................)))((((((((...))))))))........ ( -33.10) >consensus CAUCCUGAGCCAGGAUCAAACUCUCCAUAAA__AAUUAUGAUGUUUGAUUAGCUCAU_AAAUACUAA____U_U__U_UAUAAUGUUUGU___GAACUUACAAUUUAAAGUU_CCUUCGG ......((((...((((((((((................)).)))))))).))))................................................................. ( -8.46 = -9.02 + 0.56) # Strand winner: forward (0.64)

| Location | 577,882 – 577,987 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.30 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 105 + 2809422/80-200 U------------UUAGUAUUU-AUGAGCUAAUCAAACAUCAUAAUU--UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAAC .------------....(((((-.((((((((((((((.(((((...--..)))))...)))))))..))))))))))))..(((((((.(((((.......)))))..))).))))... ( -31.00) >sy_sa.0 210243 120 - 2516575/80-200 GAGAUAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAAC .................((((...((((((((((((((.((((((.....))))))...)))))))..))))))).))))..(((((((.(((((.......)))))..))).))))... ( -32.40) >sp_ag.0 91093 116 + 2160267/80-200 AACAAUAAAUCUGUCAGCGAGACA-GAAAAGAGCAAAGCUCAAACUU---UUAAUGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAGAACGCUG .........((((((.....))))-)).....((..(((((((((((---((...))))))))))(((((...))))).....))).))(((((......(((......)))..))))). ( -31.90) >consensus AA_A__A_A____UUAGUAUUU_AUGAGCUAAUCAAACAUCAUAAUU__UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAAC ................................((.........................(((((.(((((...))))))))))((((((.(((((.......)))))..))).))))).. (-18.20 = -18.20 + 0.00)

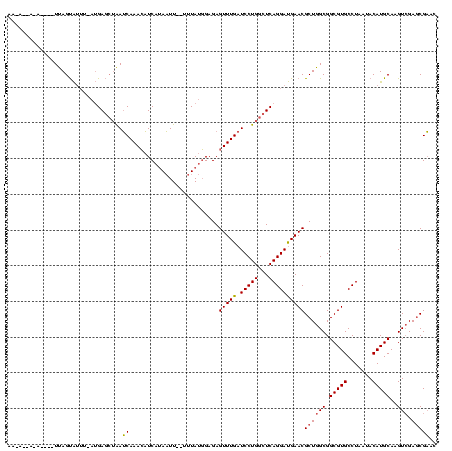
| Location | 577,882 – 578,000 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -25.20 |
| Consensus MFE | -24.12 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.29 |
| Mean z-score | 0.36 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 118 + 2809422/120-240 CCCGUCCGCCGCUAACAUCAGAGAAGCAAGCUUCUCGUCCGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA--AAUUAUG ...(..(((.((.(((....((((((....))))))....))).)).................(((.....))).)))..)((((((...))))))...............--....... ( -27.00) >sy_sa.0 210243 120 - 2516575/120-240 CCCGUCCGCCGCUAACGUCAAAGGAGCAAGCUCCUUAUCUGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAAUGAAUUAUG ...(..(((.((.((((...((((((....))))))...)))).)).................(((.....))).)))..)((((((...)))))).........(((((.....))))) ( -27.50) >sp_ag.0 91093 117 + 2160267/120-240 CCCGUUCGCAACUCAUCAGUCUAGUGUAAACACCAAACCUCAGCGUUCUACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCGUCCUGAGCCAGGAUCAAACUCUCAUUAA---AAGUUUG ..((..(((..............(((....))).........(((.......)))........(((.....))).)))..))(((((...))))).((((((........---.)))))) ( -21.10) >consensus CCCGUCCGCCGCUAACAUCAAAGGAGCAAGCUCCUAAUCUGUUCGCUCGACUUGCAUGUAUUAGGCACGCCGCCAGCGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA__AAUUAUG ...(..(((.((.(((....((((((....))))))....))).)).................(((.....))).)))..)((((((...))))))........................ (-24.12 = -22.80 + -1.32) # Strand winner: forward (1.00)

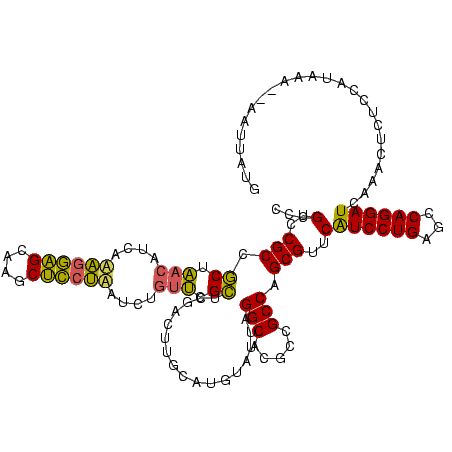
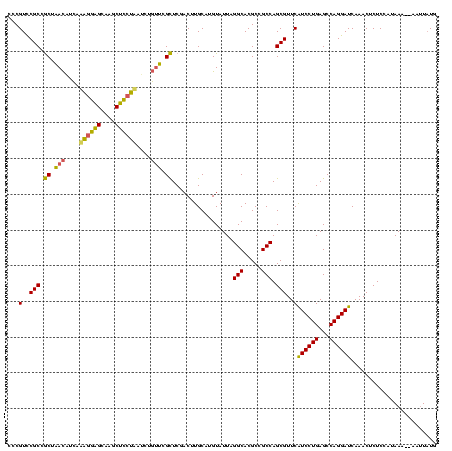
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -37.13 |
| Consensus MFE | -31.89 |
| Energy contribution | -27.13 |
| Covariance contribution | -4.76 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/200-320 GGACGAGAAGCUUGCUUCUCUGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAAC ...(((((.(((..((..((((.((....)).))))..))..))).....((((.....))))........((((..((.((((((....)))))).))...))))......)))))... ( -34.40) >sy_sa.0 210243 120 - 2516575/200-320 AGAUAAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAAC ......(..(((..(((.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...)))).............. ( -37.30) >sp_ag.0 91093 120 + 2160267/200-320 AGGUUUGGUGUUUACACUAGACUGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAA .(((((((((....)))))))))....((((((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....))))........)))))).. ( -39.70) >consensus AGAUAAGGAGCUUGCUCCUCUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUAGAAC ....((((((....))))))......(((..........(((.....)))((((.....))))))).....((((..((.((((((....)))))).))...)))).............. (-31.89 = -27.13 + -4.76)

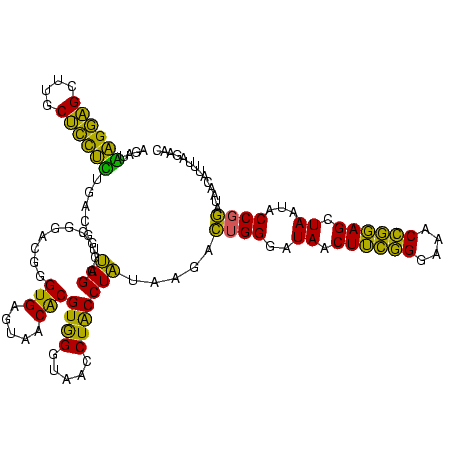
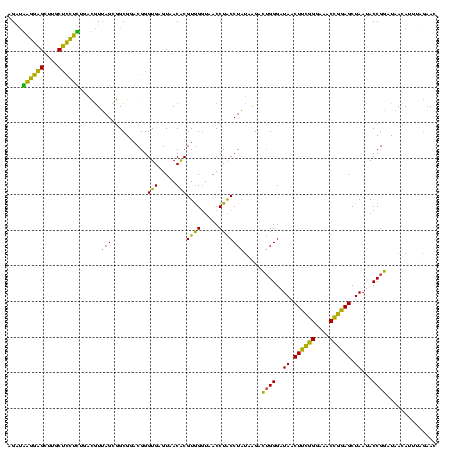
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -39.77 |
| Consensus MFE | -35.81 |
| Energy contribution | -30.50 |
| Covariance contribution | -5.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.89 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/240-360 UGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUA (((((..(((((((.....))))........((((..((.((((((....)))))).))...))))......(((((((((....))))))))))))...(((((.....)))))))))) ( -40.50) >sy_sa.0 210243 120 - 2516575/240-360 UGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUA (((((..((((((((((...)))))))....((((..((.((((((....)))))).))...))))......(((((((((....))))))))))))...(((((.....)))))))))) ( -42.40) >sp_ag.0 91093 119 + 2160267/240-360 UGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU-UCACUGU ((((......(((((...)))))))))..((((....((.((((((....)))))).))...))))....(((((((((((....)))))))))))....(((((....)))-))..... ( -36.40) >consensus UGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUAGAACCGCAUGGUUCUAAAGUGAAAGAAGGUAUUGCU_UCACUUA ..........((((.....))))...((((.((((..((.((((((....)))))).))...))))......(((((((((....)))))))))......(((((.....))))).)))) (-35.81 = -30.50 + -5.31)

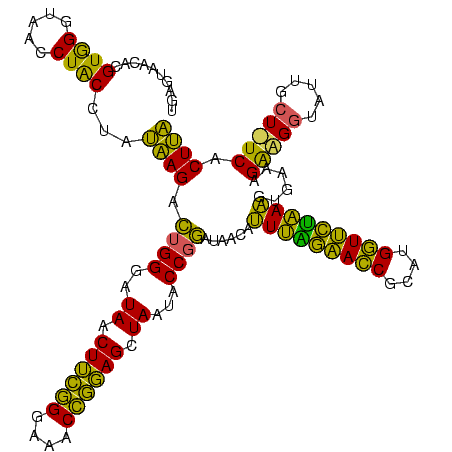
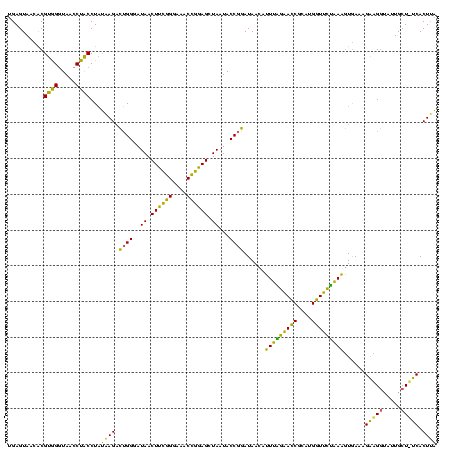
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -33.91 |
| Energy contribution | -29.60 |
| Covariance contribution | -4.31 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/280-400 CUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGUAAC ((((((....))))))....((((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))..(((((....)).)))......)))).. ( -39.10) >sy_sa.0 210243 120 - 2516575/280-400 CUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAAC ((((((....))))))....((((...((((..((((((((....))))))))(((.....(((((....(((((.......)))))......)))))....))).))))....)))).. ( -37.90) >sp_ag.0 91093 119 + 2160267/280-400 CUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU-UCACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGAGGUAAA ...(((....)))((((((((((((((...(((((((((((....)))))))))))....(((((....)))-))...............))))..)))))))))).............. ( -34.90) >consensus CUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUAGAACCGCAUGGUUCUAAAGUGAAAGAAGGUAUUGCU_UCACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUAAGGUAAC ((((((....))))))....((((........(((((((((....)))))))))......(((((.....)))))............(((.((...((....))..)).)))..)))).. (-33.91 = -29.60 + -4.31)

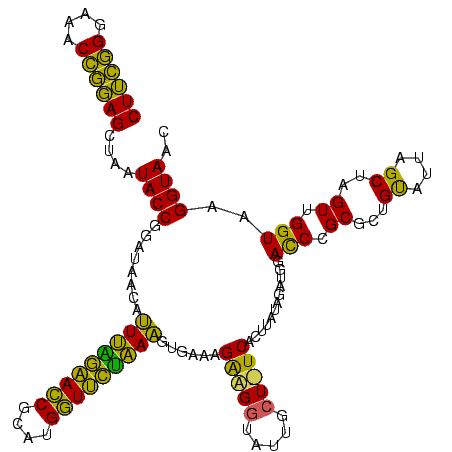
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -20.25 |
| Energy contribution | -19.03 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/280-400 GUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAG ...............(((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....)))))))))))))).(((....)))... ( -31.40) >sy_sa.0 210243 120 - 2516575/280-400 GUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCCGAAG ...............(((((((((.(.((((((...((((.((((((.((.........)).))))))))))))).)))(((..(....)..)))).))))))))).(((....)))... ( -28.10) >sp_ag.0 91093 119 + 2160267/280-400 UUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGA-AGCAAUUGCUCCUUUUAAAUAACUAACAUGUGUUAAUUACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAG ..............((((((((((..((((((......))..(((((-(((....))).........((((........)))).)))))....))))))))))))))............. ( -20.80) >consensus GUUACCUUACCAACUAGCUAAUACAGCGCGGGUCCAUCUAUAAGUGA_AGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAG ...............(((((((((...((...(((........).))..))................((((((((....))))))))..........))))))))).(((....)))... (-20.25 = -19.03 + -1.21) # Strand winner: forward (1.00)

| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -43.03 |
| Consensus MFE | -40.38 |
| Energy contribution | -39.17 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/360-480 UAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUA ..(((...((((((((((..((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...))..)))))).))))))). ( -43.00) >sy_sa.0 210243 120 - 2516575/360-480 UAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUA ..(((...(((((((((((...........((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..))))))))).))))))))). ( -44.50) >sp_ag.0 91093 120 + 2160267/360-480 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUC ...(((..((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).)))))).))) ( -41.60) >consensus UAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACAGCGCGGGUCCAUCUA ..(((...((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).))))))))). (-40.38 = -39.17 + -1.21) # Strand winner: forward (0.73)

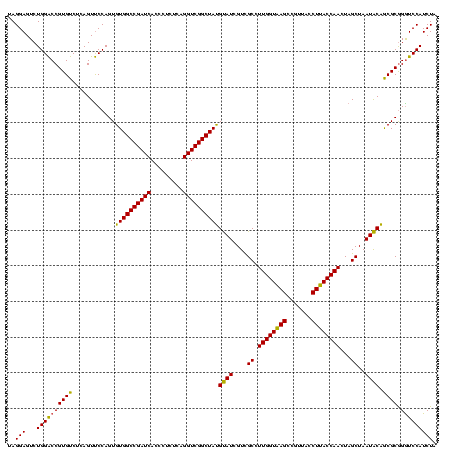
| Location | 577,882 – 578,001 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -30.40 |
| Energy contribution | -27.97 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 119 + 2809422/560-680 AUCGUAAAACUCUGUUAUUAGGGAAGAACAUAUGUGUAAGUA-ACUGUGCACAUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((.(((((((....-....)))))))..))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... ( -30.60) >sy_sa.0 210243 119 - 2516575/560-680 CUCGUAAAACUCUGUUAUUAGGGAAGAACAAACGUGUAAGUA-ACUGUGCACGUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ((((((....((((..((((((......((((((((((....-....))))))).)))......))))))))))..(((.(((((..............))))).)))..)))).))... ( -33.24) >sp_ag.0 91093 120 + 2160267/560-680 AUCGUAAAGCUCUGUUGUUAGAGAAGAACGUUGGUAGGAGUGGAAAAUCUACCAAGUGACGGUAACUAACCAGAAAGGGACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUC ........((((((..(((((......((.((((((((..(....).))))))))))........)))))))))..((.(((........))).)).)).((((((......))).))). ( -30.24) >consensus AUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGUA_ACUGUGCACAUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG .((((.....((((..((((((......((((((((((.........))))))).)))......)))))))))).....))))....(((((((((.((....)))))...))))))... (-30.40 = -27.97 + -2.43)



| Location | 577,882 – 578,001 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -31.13 |
| Energy contribution | -30.58 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 119 + 2809422/560-680 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGAUUAGGUACCGUCAAGAUGUGCACAGU-UACUUACACAUAUGUUCUUCCCUAAUAACAGAGUUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((......((..(((((......-......))))).))......))))))..)))))...)))).. ( -31.80) >sy_sa.0 210243 119 - 2516575/560-680 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGAUUAGGUACCGUCAAGACGUGCACAGU-UACUUACACGUUUGUUCUUCCCUAAUAACAGAGUUUUACGAG ......((((...((((((((((((.....))).)))))))))..(((((((((((........(((((((......-......))))))).......))))))..)))))...)))).. ( -35.06) >sp_ag.0 91093 120 + 2160267/560-680 GACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUCCCUUUCUGGUUAGUUACCGUCACUUGGUAGAUUUUCCACUCCUACCAACGUUCUUCUCUAACAACAGAGCUUUACGAU ......((((.....(((((.((((((.....)))))).)))....(((((((((........(((((((((...........))))))).))......)))))..))))))..)))).. ( -28.84) >consensus CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGAUUAGGUACCGUCAAGACGUGCACAGU_UACUUACACAUAUGUUCUUCCCUAAUAACAGAGUUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((........(((((((.............))))))).......))))))..)))))...)))).. (-31.13 = -30.58 + -0.55) # Strand winner: forward (0.68)

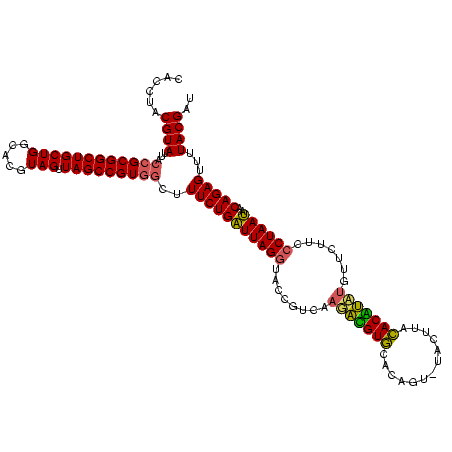

| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -37.57 |
| Consensus MFE | -34.71 |
| Energy contribution | -32.17 |
| Covariance contribution | -2.54 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/680-800 GCAAGCGUUAUCCGGAAUUAUUGGGCGUAAAGCGCGCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAA (((..((((.(((((....((..(((..(((((.(((....))))))))....)))..))((((....((((((.....))))))....))))......))))).)))..)..))).... ( -38.40) >sy_sa.0 210243 120 - 2516575/680-800 GCAAGCGUUAUCCGGAAUUAUUGGGCGUAAAGCGCGCGUAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAA ((..((((...((((.....))))(((.....)))))))..))(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))........... ( -36.00) >sp_ag.0 91093 119 + 2160267/680-800 CCGAGCGUUGUCCGGAUUUAUUGGGCGUAAAGCGAGCGCAGGCGGUUCUUUAAGUCUGAAGUUAAAGGCAGUGGCUUAACCAUUGUACG-CUUUGGAAACUGGAGGACUUGAGUGCAGAA ....((..(((((((.....)))))))....))..((((....(((((((((..((..((((.....(((((((.....)))))))..)-)))..))...)))))))))...)))).... ( -38.30) >consensus GCAAGCGUUAUCCGGAAUUAUUGGGCGUAAAGCGCGCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAA ....((((...((((.....))))((.....))))))(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..))).... (-34.71 = -32.17 + -2.54)

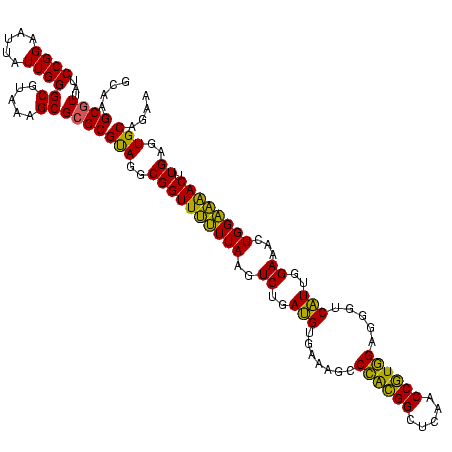
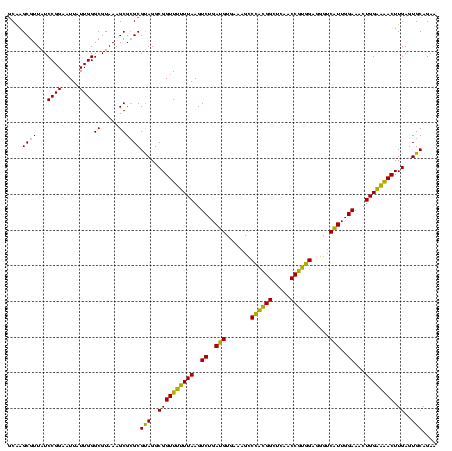
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -33.54 |
| Energy contribution | -30.67 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.88 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/720-840 GGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAG ...(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...(((((...(..(...((((....))))...)..).......))))).. ( -32.70) >sy_sa.0 210243 120 - 2516575/720-840 GGCGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAG ...(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...(((((...(..(...((((....))))...)..).......))))).. ( -35.00) >sp_ag.0 91093 119 + 2160267/720-840 GGCGGUUCUUUAAGUCUGAAGUUAAAGGCAGUGGCUUAACCAUUGUACG-CUUUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAG .(((((((((((..((..((((.....(((((((.....)))))))..)-)))..))...)))))))))...(..((....((((........)))).))..).)).............. ( -32.50) >consensus GGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAG ...(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))....(((...........((((....)))).....(((......)))))). (-33.54 = -30.67 + -2.88)

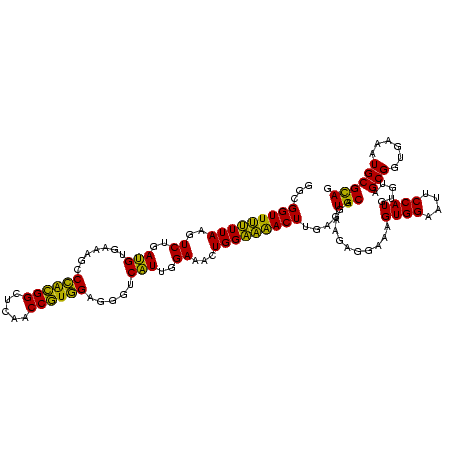
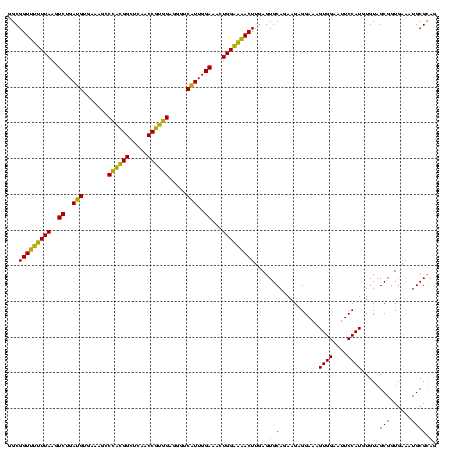
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -35.80 |
| Consensus MFE | -29.92 |
| Energy contribution | -28.60 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/760-880 CGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ...((((((((.......(((((....(((........)))......((....((((((((.(.(((........)))..).))))))))..)).))))).((....))))))))))... ( -34.00) >sy_sa.0 210243 120 - 2516575/760-880 CGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ..(.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).)))))))).. ( -35.70) >sp_ag.0 91093 119 + 2160267/760-880 CAUUGUACG-CUUUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGU ..((((((.-(((((....)..))))......))))))....((((((((...((((((((((.(((......)))....))))))))))...)))..((.((....)).)))))))... ( -37.70) >consensus CGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ..................((..((((.....................(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).))))))..)) (-29.92 = -28.60 + -1.32)

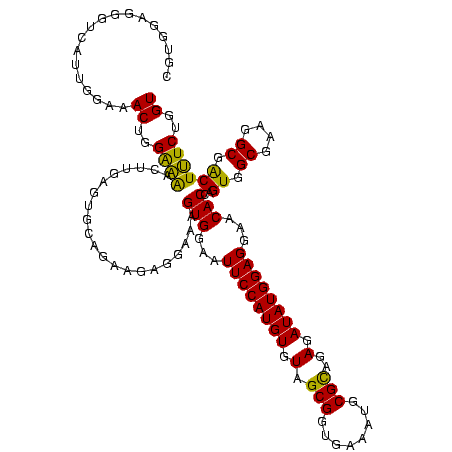
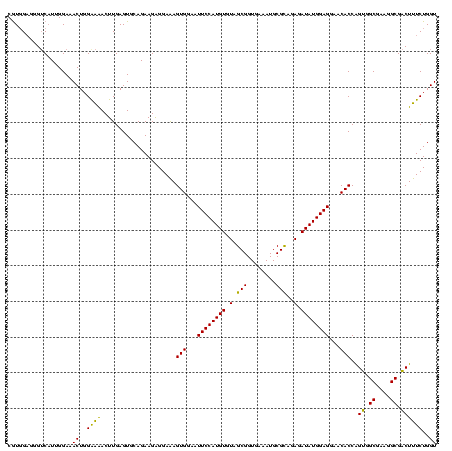
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -39.00 |
| Consensus MFE | -36.05 |
| Energy contribution | -35.17 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/800-920 GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAAC ..((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))(((((.((.((..(((......)))..))..)).))))).... ( -36.90) >sy_sa.0 210243 120 - 2516575/800-920 GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAAC ..((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))(((((.((.((..(((......)))..))..)).))))).... ( -36.90) >sp_ag.0 91093 120 + 2160267/800-920 GGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCGAAAGCGUGGGGAGCAAAC ..((((((((...((((((((((.(((......)))....))))))))))...)))..((.((....)).))))))).((.(((..((.(((((..........))))).))..))).)) ( -43.20) >consensus GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAAC ..((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).))))))).((.((...((.(((((..........))))).))...)).)) (-36.05 = -35.17 + -0.88)

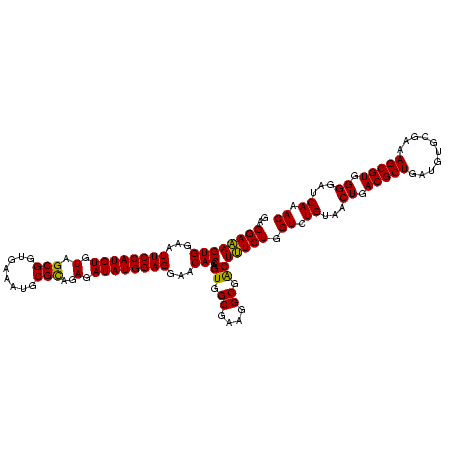
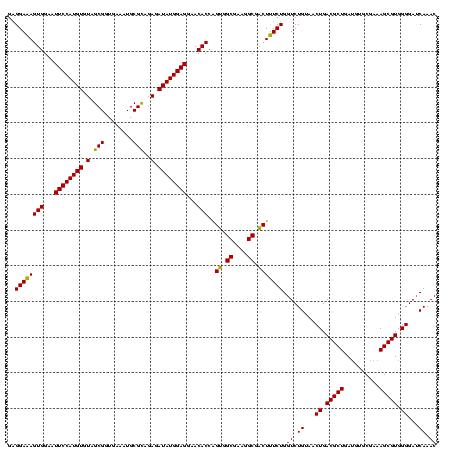
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -44.07 |
| Consensus MFE | -43.84 |
| Energy contribution | -44.07 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/920-1040 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAG .((.((..((.(((((((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).)))).))..))))..... ( -43.30) >sy_sa.0 210243 120 - 2516575/920-1040 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAG .((.((..((.(((((((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).)))).))..))))..... ( -43.30) >sp_ag.0 91093 120 + 2160267/920-1040 AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAG .((.((..((.(((((((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).)))).))..))))..... ( -45.60) >consensus AGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAG .((.((..((.(((((((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).)))).))..))))..... (-43.84 = -44.07 + 0.23)

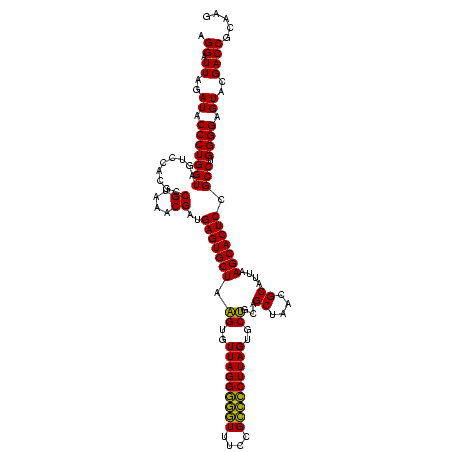
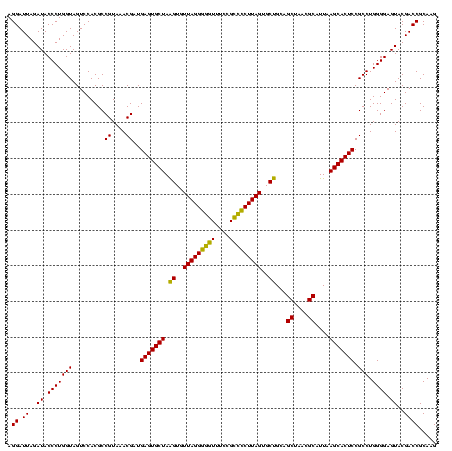
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -52.73 |
| Consensus MFE | -53.07 |
| Energy contribution | -51.30 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.27 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.69 |
| SVM RNA-class probability | 0.996411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/960-1080 UGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGG ((((.((..(((((((((....)))))))))..))...((....))....))))((((((((...((((.(((((....)))))..))))..)))......)))))..((((....)))) ( -51.90) >sy_sa.0 210243 120 - 2516575/960-1080 UGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGG ((((.((..(((((((((....)))))))))..))...((....))....))))((((((((...((((.(((((....)))))..))))..)))......)))))..((((....)))) ( -51.90) >sp_ag.0 91093 120 + 2160267/960-1080 UGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG ((((.((..(((((((((....)))))))))..))...((....))....))))((((((((...((((.(((((....)))))..))))..)))......)))))..((((....)))) ( -54.40) >consensus UGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGG .(((.((..(((((((((....)))))))))..))..)))....((.....)).((((((((...((((.(((((....)))))..))))..)))......)))))..((((....)))) (-53.07 = -51.30 + -1.77)

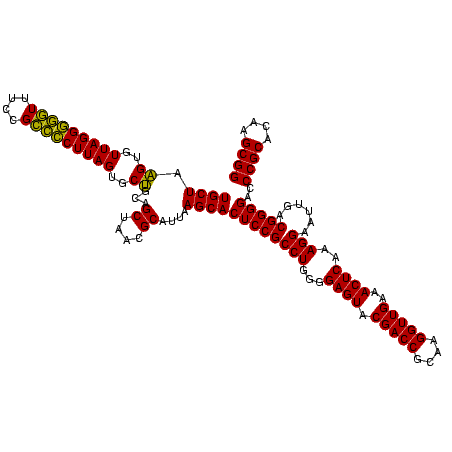
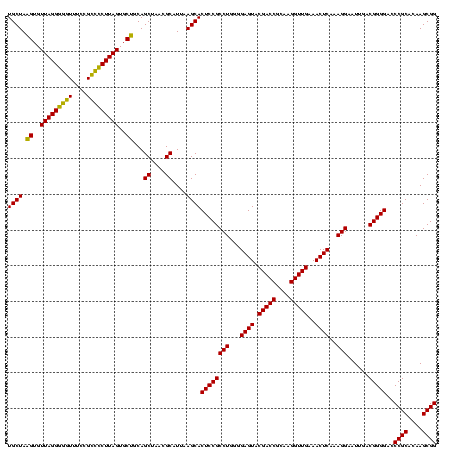
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -43.73 |
| Consensus MFE | -40.25 |
| Energy contribution | -38.70 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/960-1080 CCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCA (((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...)))))).(((((.(....))))))........... ( -43.90) >sy_sa.0 210243 120 - 2516575/960-1080 CCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCA (((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...)))))).(((((.(....))))))........... ( -43.90) >sp_ag.0 91093 120 + 2160267/960-1080 CCGCUUGUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCA ..(((.(((.(((((((((...........((((..(.(((....))).).))))....(((..((((((....((....))...))))))..))).)))...))))))..)))..))). ( -43.40) >consensus CCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCA (((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...))))))..((((......))))............. (-40.25 = -38.70 + -1.55) # Strand winner: forward (0.99)

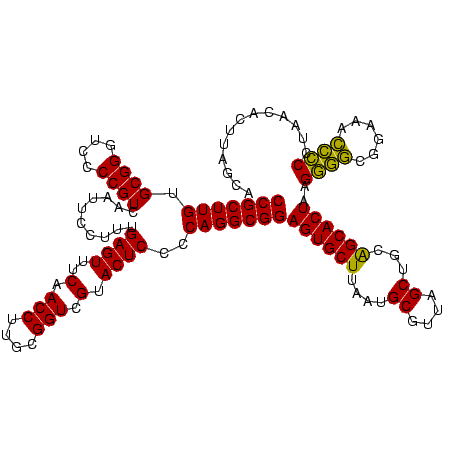
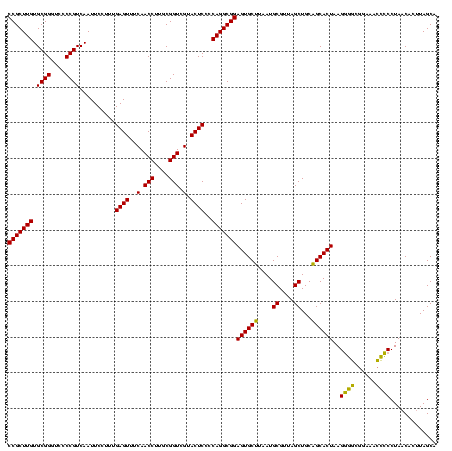
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -38.70 |
| Energy contribution | -38.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/1000-1120 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....)).(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....))))))))).....(((((..((((........))))))))).. ( -38.80) >sy_sa.0 210243 120 - 2516575/1000-1120 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....)).(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....))))))))).....(((((..((((........))))))))).. ( -38.80) >sp_ag.0 91093 120 + 2160267/1000-1120 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -39.80) >consensus UAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....)).(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....))))))))).....(((((..((((........))))))))).. (-38.70 = -38.70 + -0.00)

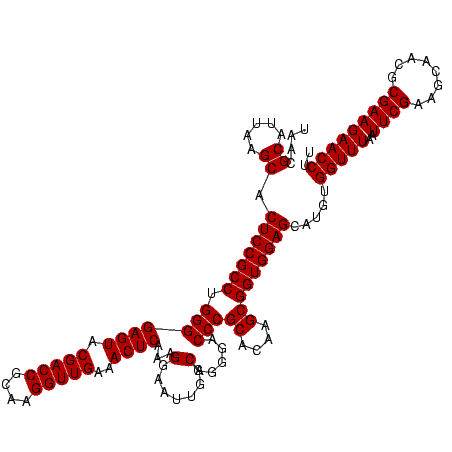
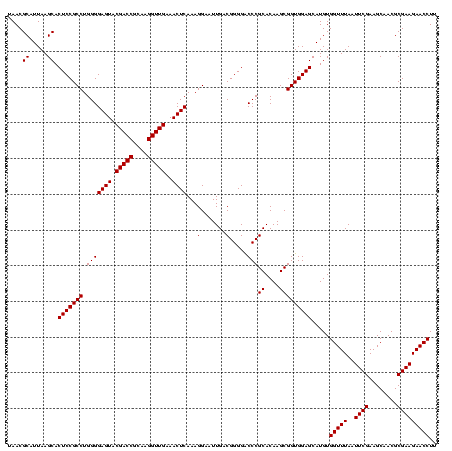
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -40.35 |
| Energy contribution | -41.80 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/1200-1320 UGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAA ....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....)))))..... ( -42.20) >sy_sa.0 210243 120 - 2516575/1200-1320 UGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGAA ....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....)))))..... ( -41.90) >sp_ag.0 91093 120 + 2160267/1200-1320 UGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAA ....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....)))))..... ( -41.30) >consensus UGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGAA ....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....)))))..... (-40.35 = -41.80 + 1.45)

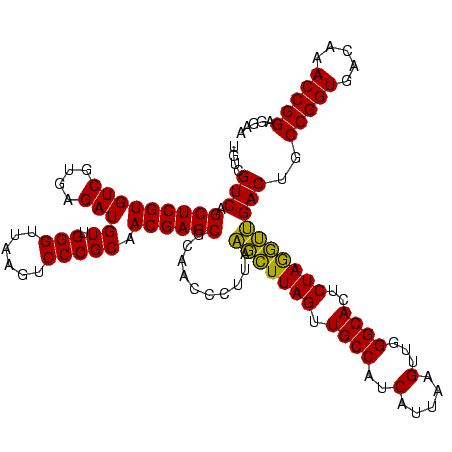
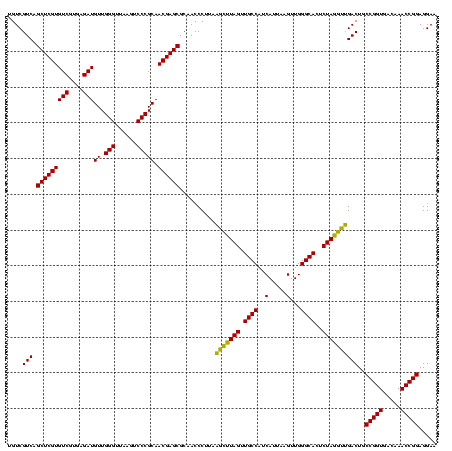
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -33.30 |
| Energy contribution | -31.20 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/1320-1440 GGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAG ..((((((..(((((..........((((..........))))......(((......))))))........(((..((...(((....)))...))....))).....))..)))))). ( -32.70) >sy_sa.0 210243 120 - 2516575/1320-1440 GGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCUAAACCGCGAGGUCAUGCAAAUCCCAUAAAGUUGUUCUCAG ..((((((..(((((..........((((..........))))......(((......))))))........(((..((...(((....)))...))....))).....))..)))))). ( -32.30) >sp_ag.0 91093 120 + 2160267/1320-1440 GGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCUACACACGUGCUACAAUGGUUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAAUCUCAG ...((((((((.(......).)))).))))...(((....((((......((((((.......))))))...(((..((..((((....))))..))....)))....))))....))). ( -38.00) >consensus GGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCAAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAG ...((((((((.(......).)))).))))...(((....(((.((....))))).....(((((((.....(((..((...(((....)))...))....))).....)))))))))). (-33.30 = -31.20 + -2.10)

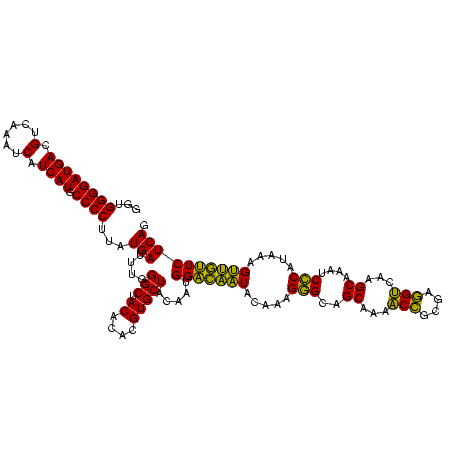
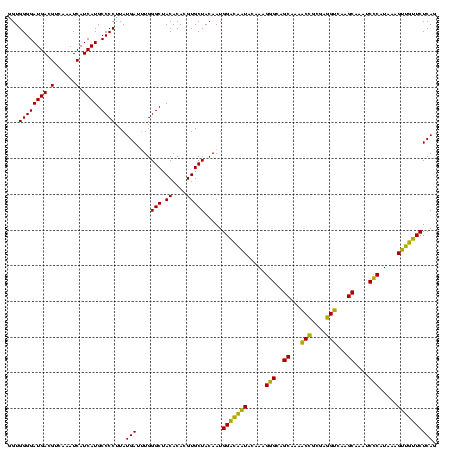
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -34.72 |
| Energy contribution | -31.40 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.27 |
| Structure conservation index | 1.04 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/1320-1440 CUGAGAACAACUUUAUGGGAUUUGCUUGACCUCGCGGUUUCGCUGCCCUUUGUAUUGUCCAUUGUAGCACGUGUGUAGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACC ...............((((....((..((((....))))..))(((((((((((((.......((.(((....))).))...))).)))))))))).((((((.....)))))))))).. ( -31.10) >sy_sa.0 210243 120 - 2516575/1320-1440 CUGAGAACAACUUUAUGGGAUUUGCAUGACCUCGCGGUUUAGCUGCCCUUUGUAUUGUCCAUUGUAGCACGUGUGUAGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACC ...............((((...(((((((((....)))...(((((....((.......))..))))).))))))...))))........((((.((((((......))))))))))... ( -33.40) >sp_ag.0 91093 120 + 2160267/1320-1440 CUGAGAUUGGCUUUAAGAGAUUAGCUUGCCGUCACCGGCUUGCGACUCGUUGUACCAACCAUUGUAGCACGUGUGUAGCCCAGGUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACC ....((((((((...........((..((((....))))..)).((.(((.((..((.....))..))))).))..))))..))))....((((.((((((......))))))))))... ( -35.40) >consensus CUGAGAACAACUUUAUGGGAUUUGCUUGACCUCGCGGUUUAGCUGCCCUUUGUAUUGUCCAUUGUAGCACGUGUGUAGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACC .((((.((((......(((....((..((((....))))..))..)))......)))).)...((.(((....))).)).....)))...((((.((((((......))))))))))... (-34.72 = -31.40 + -3.32) # Strand winner: reverse (0.67)

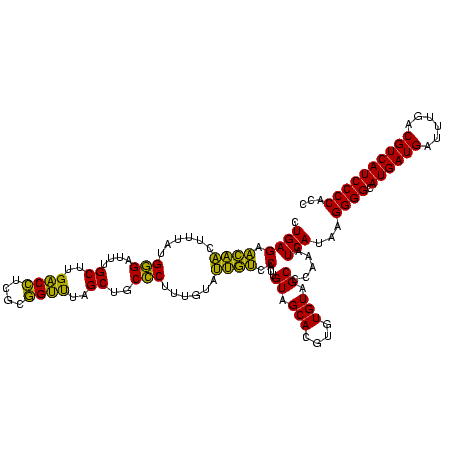

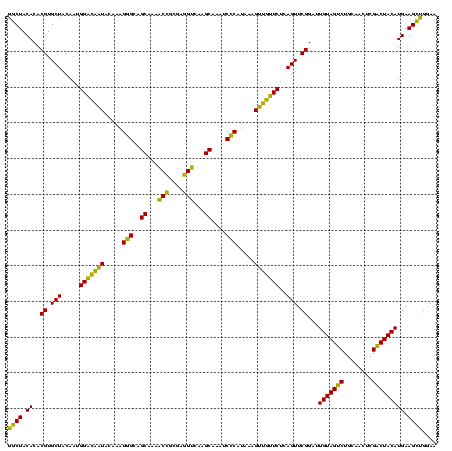
| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -37.12 |
| Energy contribution | -34.13 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.05 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/1360-1480 GGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAA ((((.((..((.(((.....(((((((.....(((..((...(((....)))...))....))).....)))))))..))).))...(((((((........))))))))).)))).... ( -33.10) >sy_sa.0 210243 120 - 2516575/1360-1480 GGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCUAAACCGCGAGGUCAUGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAA ((((.((..((.(((.....(((((((.....(((..((...(((....)))...))....))).....)))))))..))).))...(((((((........))))))))).)))).... ( -32.70) >sp_ag.0 91093 120 + 2160267/1360-1480 GGCUACACACGUGCUACAAUGGUUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAAUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGUCGGAA ((((.((..((.(((.....(((((((.....(((..((..((((....))))..))....))).....)))))))..))).))...(((((((........))))))))).)))).... ( -40.70) >consensus GGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCAAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAA ((((.((..((.(((.....(((((((.....(((..((...(((....)))...))....))).....)))))))..))).))...(((((((........))))))))).)))).... (-37.12 = -34.13 + -2.99)


| Location | 577,882 – 578,002 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -31.29 |
| Energy contribution | -27.63 |
| Covariance contribution | -3.65 |
| Combinations/Pair | 1.30 |
| Mean z-score | 0.82 |
| Structure conservation index | 1.09 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 120 + 2809422/1360-1480 UUCCAGCUUCAUGUAGUCGAGUUGCAGACUACAAUCCGAACUGAGAACAACUUUAUGGGAUUUGCUUGACCUCGCGGUUUCGCUGCCCUUUGUAUUGUCCAUUGUAGCACGUGUGUAGCC .(((((.(((.(((((((........)))))))....)))))).))((((..(...(((....((..((((....))))..))..)))...)..)))).....((.(((....))).)). ( -27.90) >sy_sa.0 210243 120 - 2516575/1360-1480 UUCCAGCUUCAUGUAGUCGAGUUGCAGACUACAAUCCGAACUGAGAACAACUUUAUGGGAUUUGCAUGACCUCGCGGUUUAGCUGCCCUUUGUAUUGUCCAUUGUAGCACGUGUGUAGCC .(((((.(((.(((((((........)))))))....)))))).))((((..(...(((....((..((((....))))..))..)))...)..)))).....((.(((....))).)). ( -26.90) >sp_ag.0 91093 120 + 2160267/1360-1480 UUCCGACUUCAUGUAGGCGAGUUGCAGCCUACAAUCCGAACUGAGAUUGGCUUUAAGAGAUUAGCUUGCCGUCACCGGCUUGCGACUCGUUGUACCAACCAUUGUAGCACGUGUGUAGCC .....((..(((((.((((((((((((((..(((((........)))))(((..........)))...........))).)))))))))))((..((.....))..))))))).)).... ( -31.50) >consensus UUCCAGCUUCAUGUAGUCGAGUUGCAGACUACAAUCCGAACUGAGAACAACUUUAUGGGAUUUGCUUGACCUCGCGGUUUAGCUGCCCUUUGUAUUGUCCAUUGUAGCACGUGUGUAGCC .(((((.(((.(((((((........)))))))....)))))).))((((......(((....((..((((....))))..))..)))......)))).....((.(((....))).)). (-31.29 = -27.63 + -3.65) # Strand winner: forward (0.97)


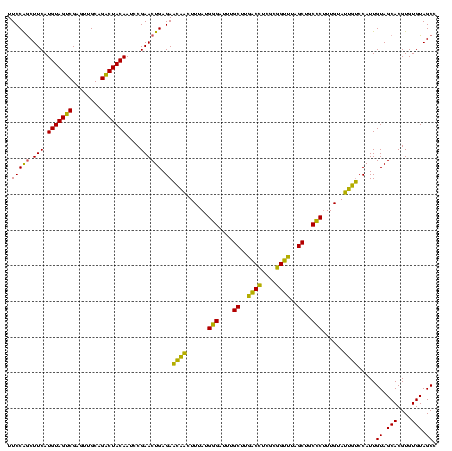
| Location | 577,882 – 578,001 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.43 |
| Mean single sequence MFE | -46.47 |
| Consensus MFE | -46.02 |
| Energy contribution | -45.13 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 119 + 2809422/1560-1680 UUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUC ((((..(((((((...((((..(((..((.....-)).))).)))))))..))))..))))......(((((((.((....))....((((((((((....))))))))))..))))))) ( -46.40) >sy_sa.0 210243 120 - 2516575/1560-1680 UUGUAACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUC ((((..(((((((...((((..(((..(((....))).))).)))))))..))))..))))......(((((((.((....))....((((((((((....))))))))))..))))))) ( -47.10) >sp_ag.0 91093 119 + 2160267/1560-1680 UUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUC ((((..((((...((.((((..(((..((.....-)).))).)))).))..))))..))))......(((((((.((....))....((((((((((....))))))))))..))))))) ( -45.90) >consensus UUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA_GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUC ((((..((((....(.((((..(((..((......)).))).)))).)...))))..))))......(((((((.((....))....((((((((((....))))))))))..))))))) (-46.02 = -45.13 + -0.88)

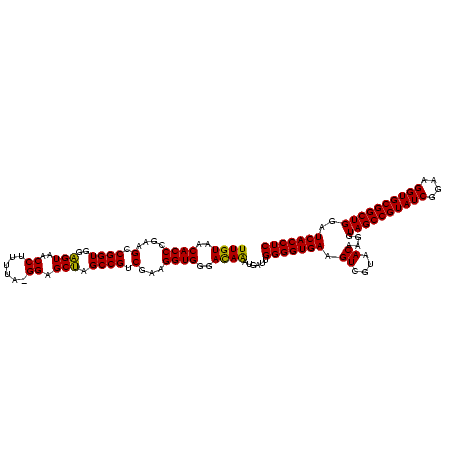

| Location | 577,882 – 577,997 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -32.80 |
| Energy contribution | -32.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 115 + 2809422/1600-1720 UAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAU--AUUCGGAACAUCU---UCUUCAGA .....(..(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((...--..)))...)))))---))..)... ( -38.90) >sy_sa.0 210243 115 - 2516575/1600-1720 UAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAU--AUUCGGAACAUCU---UCUUACGA ....(((.(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((...--..)))...)))))---))..))). ( -40.30) >sp_ag.0 91093 120 + 2160267/1600-1720 CAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAAACCUGCCAUUUGCGUCUUGUUU ...(((((...)))))((((((((..((((((((.((....))....((((((((((....))))))))))..)))))(((((((....)).)))))......)))....)))).)))). ( -43.30) >consensus UAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAU__AUUCGGAACAUCU___UCUUAAGA ..(((......))).........((..(((((((.((....))....((((((((((....))))))))))..))))).))..))................................... (-32.80 = -32.80 + 0.00)

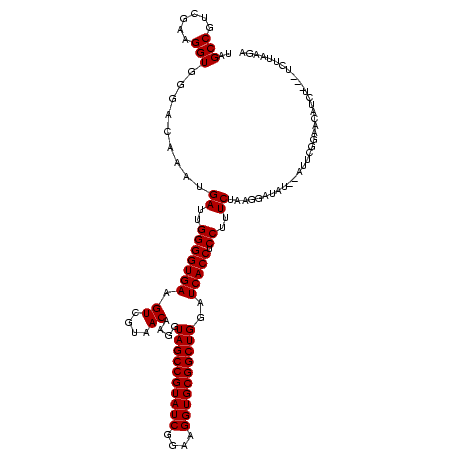
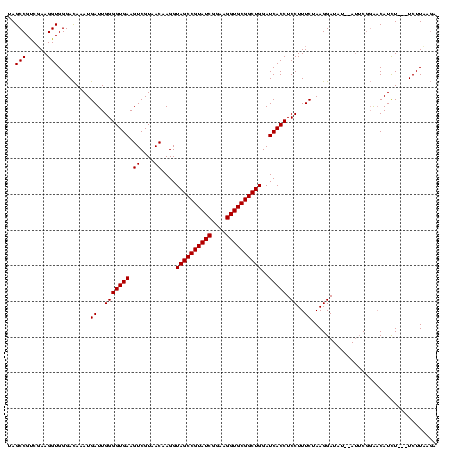
| Location | 577,882 – 577,996 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.97 |
| Mean single sequence MFE | -40.07 |
| Consensus MFE | -29.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 577882 114 + 2809422/1616-1736 GACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAU--AUUCGGAACAUCU---UCUUCAGAAGAUGC-GGAAUAAUG .......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))........(--((((.(..(((((---((....))))))))-.)))))... ( -41.70) >sy_sa.0 210243 115 - 2516575/1616-1736 GACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAU--AUUCGGAACAUCU---UCUUACGAAGAUGCAGGAAUAACA .......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))........(--((((....(((((---((....)))))))...)))))... ( -40.10) >sp_ag.0 91093 120 + 2160267/1616-1736 GAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAAACCUGCCAUUUGCGUCUUGUUUAGUUUUGAGAGGUCUU ...(((((...(((((((.((....))....((((((((((....))))))))))..)))))(((((((....)).)))))..))...)))))((.(((((........))))).))... ( -38.40) >consensus GACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAU__AUUCGGAACAUCU___UCUUAAGAAGAUGC_GGAAUAAUA .......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))................................................... (-29.70 = -29.70 + -0.00)

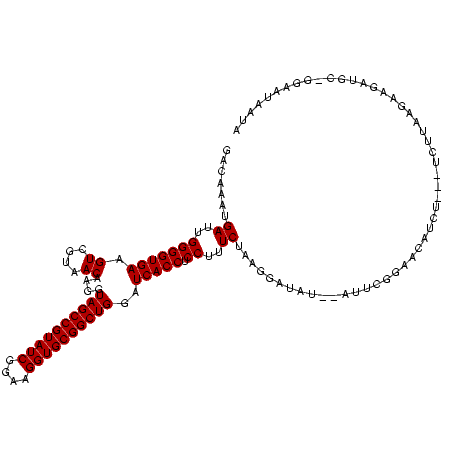
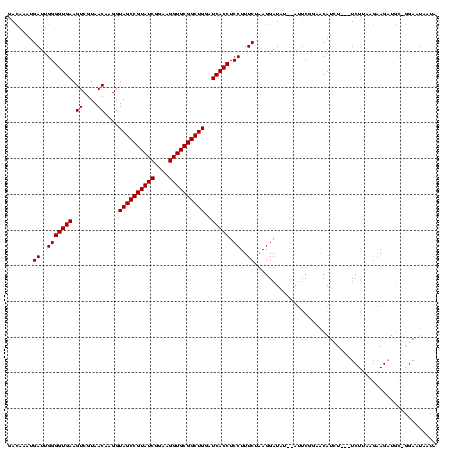
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:35 2006