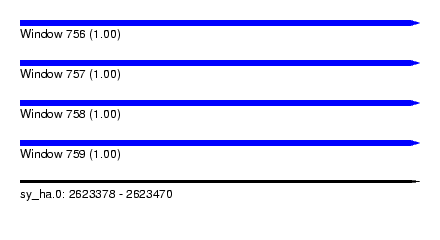
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 2,623,378 – 2,623,470 |
| Length | 92 |
| Max. P | 0.999910 |

| Location | 2,623,378 – 2,623,470 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 64.23 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
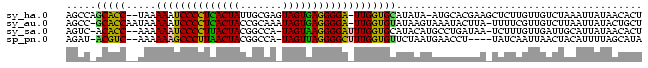
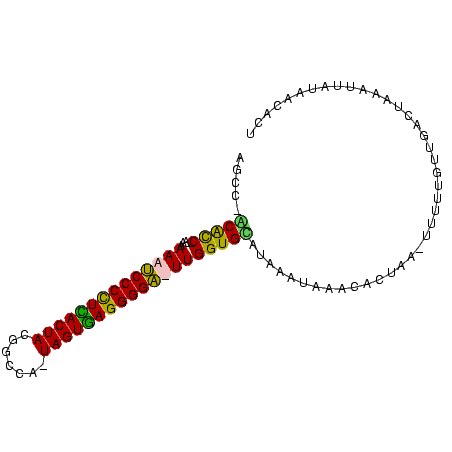
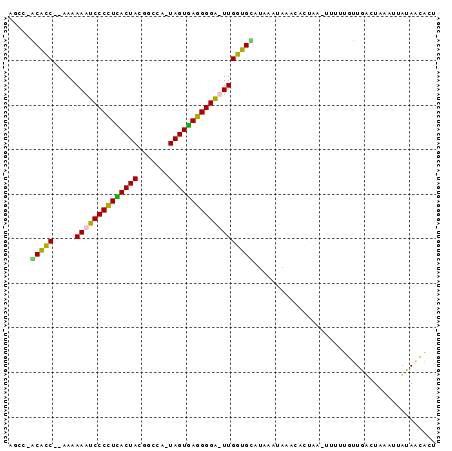
>sy_ha.0 2623378 92 + 2685015 AGUGUUAUAAUUUAGACAACAAGAGCUUCGUGCAU-UAUAUGCACCAA-UCCCCUCACUACUCGCAAUAGUGAGGGGAUUUUUA--GGUGCUGGCU ..((((........))))......((.....))..-.....(((((((-(((((((((((.......)))))))))))))....--)))))..... ( -31.50) >sy_au.0 902285 93 - 2809422 AGCAGUAUAAUUAAGACAACGAAAA-UAAGUAUUUACUUAUACACCAA-UCCCCUCACUAUUUGCGGUAGUGAGGGGAUUUUUAUUGGUGC-GGCU (((.((((...(((((........(-(((((....))))))......(-((((((((((((.....))))))))))))))))))...))))-.))) ( -32.50) >sy_sa.0 1702732 91 + 2516575 AGUGUUAUAAUGCAAUCAACAAAGA-UUAUCAGGCAUGUAUGCACCAAAUCCCCUUACUA-UGGCCGUAGUAAGGGGAUUUUUU--GGUGU-GACU ...(((...((((((((......))-)).....))))....(((((((((((((((((((-(....)))))))))))))...))--)))))-))). ( -33.20) >sp_pn.0 72987 88 - 2038615 UAUGCUAAAAUGUAGUUAAUUGAUA----AGGUUCAUUAGAACACCAAAGCCCCUAACUA-UGGCCGUAGUUAAGGGCUUUUUU--GACGU-AUCU ..(((......)))(((((......----..((((....))))...(((((((.((((((-(....))))))).))))))).))--)))..-.... ( -24.30) >consensus AGUGUUAUAAUGUAGACAACAAAAA_UUAGUAUUCAUUUAUACACCAA_UCCCCUCACUA_UGGCCGUAGUGAGGGGAUUUUUA__GGUGC_GGCU .........................................(((((((((((((((((((.......)))))))))))))).....)))))..... (-21.70 = -21.95 + 0.25)

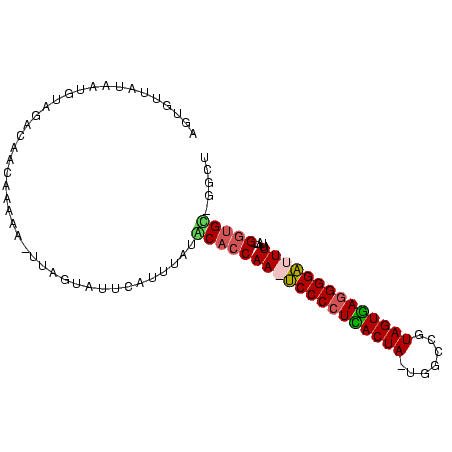
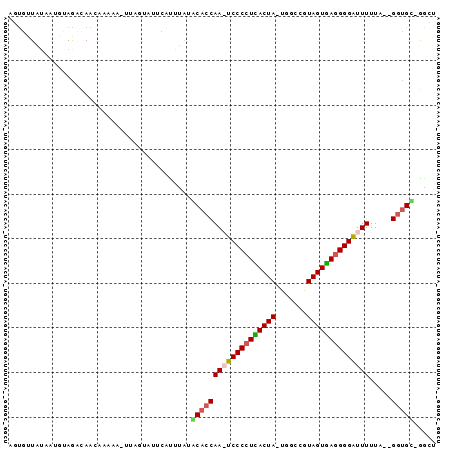
| Location | 2,623,378 – 2,623,470 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 64.23 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -24.34 |
| Energy contribution | -23.28 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.42 |
| Mean z-score | -4.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2623378 92 + 2685015 AGCCAGCACC--UAAAAAUCCCCUCACUAUUGCGAGUAGUGAGGGGA-UUGGUGCAUAUA-AUGCACGAAGCUCUUGUUGUCUAAAUUAUAACACU .....(((((--....((((((((((((((.....))))))))))))-))))))).((((-((......(((....)))......))))))..... ( -35.60) >sy_au.0 902285 93 - 2809422 AGCC-GCACCAAUAAAAAUCCCCUCACUACCGCAAAUAGUGAGGGGA-UUGGUGUAUAAGUAAAUACUUA-UUUUCGUUGUCUUAAUUAUACUGCU (((.-((((((......((((((((((((.......)))))))))))-)))))))((((((....)))))-).....................))) ( -30.60) >sy_sa.0 1702732 91 + 2516575 AGUC-ACACC--AAAAAAUCCCCUUACUACGGCCA-UAGUAAGGGGAUUUGGUGCAUACAUGCCUGAUAA-UCUUUGUUGAUUGCAUUAUAACACU .((.-.((((--((...((((((((((((......-))))))))))))))))))...)).....((.(((-((......))))))).......... ( -28.50) >sp_pn.0 72987 88 - 2038615 AGAU-ACGUC--AAAAAAGCCCUUAACUACGGCCA-UAGUUAGGGGCUUUGGUGUUCUAAUGAACCU----UAUCAAUUAACUACAUUUUAGCAUA .(((-(....--...((((((((((((((......-))))))))))))))((.((((....))))))----))))..................... ( -25.20) >consensus AGCC_ACACC__AAAAAAUCCCCUCACUACGGCCA_UAGUGAGGGGA_UUGGUGCAUAAAUAAACACUAA_UUUUUGUUGACUAAAUUAUAACACU .....(((((.....((((((((((((((.......)))))))))))))))))))......................................... (-24.34 = -23.28 + -1.06) # Strand winner: reverse (0.89)
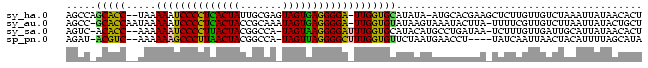
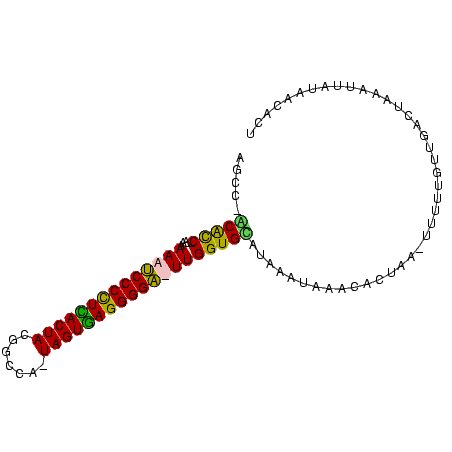
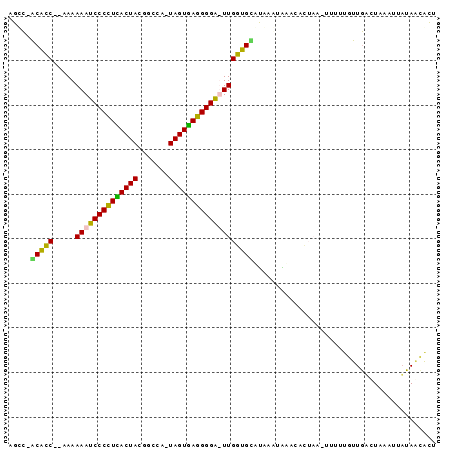
| Location | 2,623,378 – 2,623,470 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 64.23 |
| Mean single sequence MFE | -30.38 |
| Consensus MFE | -21.70 |
| Energy contribution | -21.95 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.14 |
| Structure conservation index | 0.71 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2623378 92 + 2685015/0-96 AGUGUUAUAAUUUAGACAACAAGAGCUUCGUGCAU-UAUAUGCACCAA-UCCCCUCACUACUCGCAAUAGUGAGGGGAUUUUUA--GGUGCUGGCU ..((((........))))......((.....))..-.....(((((((-(((((((((((.......)))))))))))))....--)))))..... ( -31.50) >sy_au.0 902285 93 - 2809422/0-96 AGCAGUAUAAUUAAGACAACGAAAA-UAAGUAUUUACUUAUACACCAA-UCCCCUCACUAUUUGCGGUAGUGAGGGGAUUUUUAUUGGUGC-GGCU (((.((((...(((((........(-(((((....))))))......(-((((((((((((.....))))))))))))))))))...))))-.))) ( -32.50) >sy_sa.0 1702732 91 + 2516575/0-96 AGUGUUAUAAUGCAAUCAACAAAGA-UUAUCAGGCAUGUAUGCACCAAAUCCCCUUACUA-UGGCCGUAGUAAGGGGAUUUUUU--GGUGU-GACU ...(((...((((((((......))-)).....))))....(((((((((((((((((((-(....)))))))))))))...))--)))))-))). ( -33.20) >sp_pn.0 72987 88 - 2038615/0-96 UAUGCUAAAAUGUAGUUAAUUGAUA----AGGUUCAUUAGAACACCAAAGCCCCUAACUA-UGGCCGUAGUUAAGGGCUUUUUU--GACGU-AUCU ..(((......)))(((((......----..((((....))))...(((((((.((((((-(....))))))).))))))).))--)))..-.... ( -24.30) >consensus AGUGUUAUAAUGUAGACAACAAAAA_UUAGUAUUCAUUUAUACACCAA_UCCCCUCACUA_UGGCCGUAGUGAGGGGAUUUUUA__GGUGC_GGCU .........................................(((((((((((((((((((.......)))))))))))))).....)))))..... (-21.70 = -21.95 + 0.25)

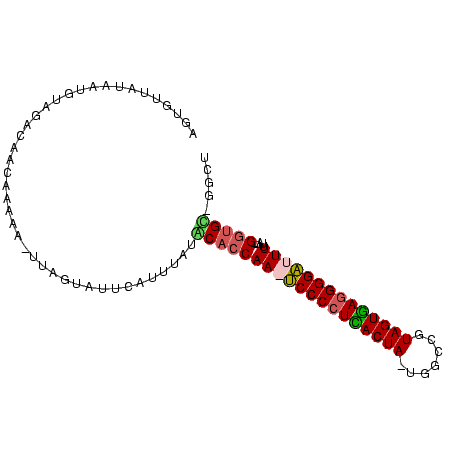
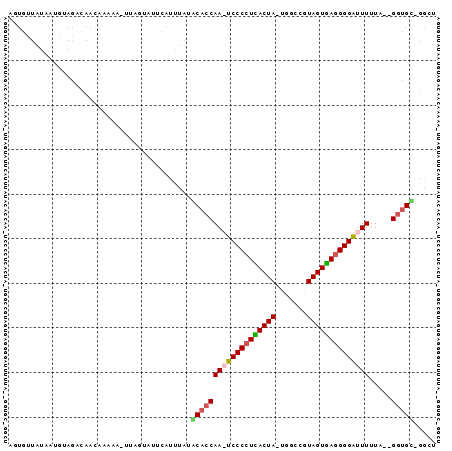
| Location | 2,623,378 – 2,623,470 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 64.23 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -24.34 |
| Energy contribution | -23.28 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.42 |
| Mean z-score | -4.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2623378 92 + 2685015/0-96 AGCCAGCACC--UAAAAAUCCCCUCACUAUUGCGAGUAGUGAGGGGA-UUGGUGCAUAUA-AUGCACGAAGCUCUUGUUGUCUAAAUUAUAACACU .....(((((--....((((((((((((((.....))))))))))))-))))))).((((-((......(((....)))......))))))..... ( -35.60) >sy_au.0 902285 93 - 2809422/0-96 AGCC-GCACCAAUAAAAAUCCCCUCACUACCGCAAAUAGUGAGGGGA-UUGGUGUAUAAGUAAAUACUUA-UUUUCGUUGUCUUAAUUAUACUGCU (((.-((((((......((((((((((((.......)))))))))))-)))))))((((((....)))))-).....................))) ( -30.60) >sy_sa.0 1702732 91 + 2516575/0-96 AGUC-ACACC--AAAAAAUCCCCUUACUACGGCCA-UAGUAAGGGGAUUUGGUGCAUACAUGCCUGAUAA-UCUUUGUUGAUUGCAUUAUAACACU .((.-.((((--((...((((((((((((......-))))))))))))))))))...)).....((.(((-((......))))))).......... ( -28.50) >sp_pn.0 72987 88 - 2038615/0-96 AGAU-ACGUC--AAAAAAGCCCUUAACUACGGCCA-UAGUUAGGGGCUUUGGUGUUCUAAUGAACCU----UAUCAAUUAACUACAUUUUAGCAUA .(((-(....--...((((((((((((((......-))))))))))))))((.((((....))))))----))))..................... ( -25.20) >consensus AGCC_ACACC__AAAAAAUCCCCUCACUACGGCCA_UAGUGAGGGGA_UUGGUGCAUAAAUAAACACUAA_UUUUUGUUGACUAAAUUAUAACACU .....(((((.....((((((((((((((.......)))))))))))))))))))......................................... (-24.34 = -23.28 + -1.06) # Strand winner: reverse (0.89)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:13 2006