
| Sequence ID | sp_pn.0 |
|---|---|
| Location | 1,854,960 – 1,855,080 |
| Length | 120 |
| Max. P | 0.999838 |

| Location | 1,854,960 – 1,855,075 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.69 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -13.11 |
| Energy contribution | -9.67 |
| Covariance contribution | -3.44 |
| Combinations/Pair | 1.68 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
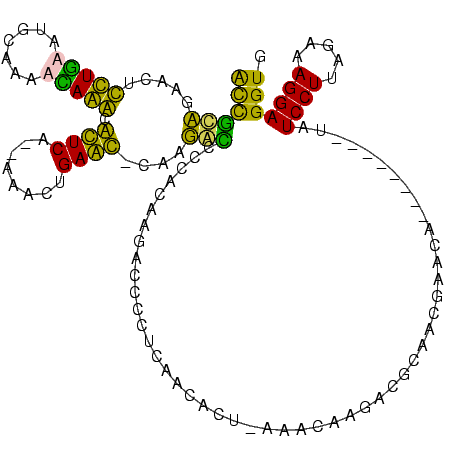
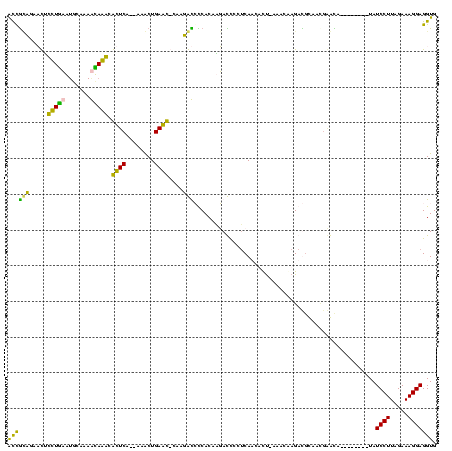
>sp_pn.0 1854960 115 + 2038615/40-160 ACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGC-UAAGGCCCCACAAGACCUCUCAAGACU-AAACAAGAC-CAAUGCGCAG--UUCCUUAUCCUUAGAAAGGAGGUG .......(((((((((.(((...))).))((((........))))-(((((........((....))..((((-...((....-...))...))--))......)))))...))))))). ( -25.30) >sp_ag.0 1908203 118 - 2160267/40-160 ACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGC-UAAGGCCCCACAAGACCUCUCAAAACU-AAACAAGACGCAAAUGGCAGGUUUCCUUAUCCUUAGAAAGGAGGUG .......(((((((((.(((...))).))((((........))))-(((((.......((((((.......((-.....))..((.....))))))))......)))))...))))))). ( -31.82) >sy_ha.0 2542316 101 + 2685015/40-160 GUUGUAGAAUGUUUAAA------AUAAACAUUCA--AAACUGAAUACAAUAUGUCACGUUAUCCCUCAUCUCU---ACGAGAUGUUCCGAAUA--------UAUCCUUAGAAAGGAGGUG (((((((((((((((..------.))))))))).--........)))))).............((((...(((---(.(.(((((.......)--------))))).))))...)))).. ( -20.50) >sy_au.0 2278565 110 - 2809422/40-160 ACCAAAAAAUAUUUGAAUGUUAAAUAAACAUUCA--AAACUGAAUACAAUAUGUCACGUUAUUCCGCAUCUUCUGAAGAAGAUGUUCCGAAUA--------UAUCCUUAGAAAGGAGGUG (((........((((((((((.....))))))))--)).....................(((((.((((((((....))))))))...)))))--------..((((.....))))))). ( -26.90) >consensus ACCGCAGAACUCCUGAAUGCAAAACAAACACUCA__AAACUGAAC_CAAGACCCCACAAGACCCCUCAACACU_AAACAAGACGCAACGAACA________UAUCCUUAGAAAGGAGGUG ((((((.....(((((.......))))).((((........))))....)))...................................................((((.....))))))). (-13.11 = -9.67 + -3.44)

| Location | 1,854,960 – 1,855,075 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.14 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -27.25 |
| Energy contribution | -28.88 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
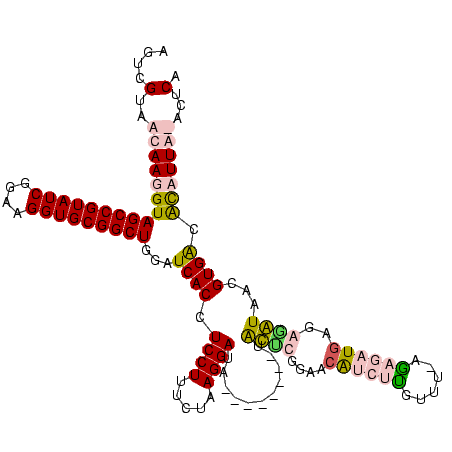
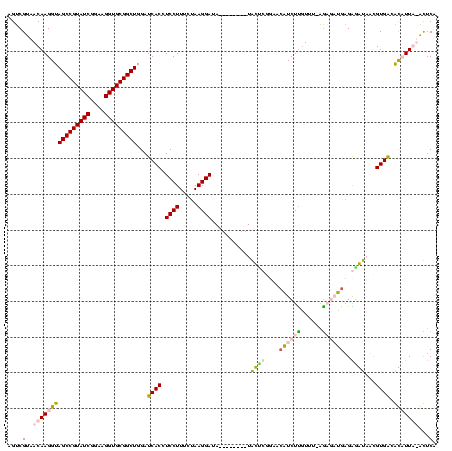
>sp_pn.0 1854960 115 + 2038615/80-200 AGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAA--CUGCGCAUUG-GUCUUGUUU-AGUCUUGAGAGGUCUUGUGGGGCCUUA-GCUCA .......((((((((((((((((....))))))))))........((((((..(((((...(((--(...((....-))...))))-.))))))))))))))))).((((....-)))). ( -39.10) >sp_ag.0 1908203 118 - 2160267/80-200 AGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAAACCUGCCAUUUGCGUCUUGUUU-AGUUUUGAGAGGUCUUGUGGGGCCUUA-GCUCA ((.(((((..(((((((((((((....)))))))))........(((((((....)).))))).)))).....))))).)).....-.....((((((((((....))))))..-.)))) ( -40.20) >sy_ha.0 2542316 109 + 2685015/80-200 AGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA--------UAUUCGGAACAUCUCGU---AGAGAUGAGGGAUAACGUGACAUAUUGUAUUCA .((((........((((((((((....)))))))))).(((.((((.((((((.((((.--------..........))))..)---))))).)))))))....))))............ ( -36.50) >sy_au.0 2278565 112 - 2809422/80-200 AGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA--------UAUUCGGAACAUCUUCUUCAGAAGAUGCGGAAUAACGUGACAUAUUGUAUUCA .......((((..((((((((((....))))))))))..((((.((((.....))))..--------(((((.(..(((((((....)))))))).)))))..))))....))))..... ( -38.60) >consensus AGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA________UACUCGGAACAUCUUGUUU_AGAGAUGAGAGAUAACGUGACACAUUA_ACUCA ....(..((((((((((((((((....)))))))))...((((.((((.....))))...........((((....((((((......))))))..))))...)))).)))))))...). (-27.25 = -28.88 + 1.62) # Strand winner: reverse (1.00)

| Location | 1,854,960 – 1,855,077 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -35.15 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 117 + 2038615/120-240 AGGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAA--CUGCGCAUUG-GUC ....((((((((((...)))))..((((.....(((((((.((....))....((((((((((....))))))))))..)))))))...))))...........--))).))....-... ( -41.70) >sp_ag.0 1908203 120 - 2160267/120-240 AGGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAAACCUGCCAUUUGCGUC ....((...(((((...)))))...(((((...(((((((.((....))....((((((((((....))))))))))..)))))(((((((....)).)))))..))...)))))))... ( -41.90) >sy_ha.0 2542316 111 + 2685015/120-240 -GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA--------UAUUCGGAACAUC -((......)).((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))..--------..)))))...... ( -36.80) >sy_au.0 2278565 112 - 2809422/120-240 AGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA--------UAUUCGGAACAUC .((......)).((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))..--------..)))))...... ( -37.00) >consensus AGGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA________UACUCGGAACAUC .....((.(((......))).))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))............................. (-35.15 = -34.90 + -0.25) # Strand winner: reverse (1.00)

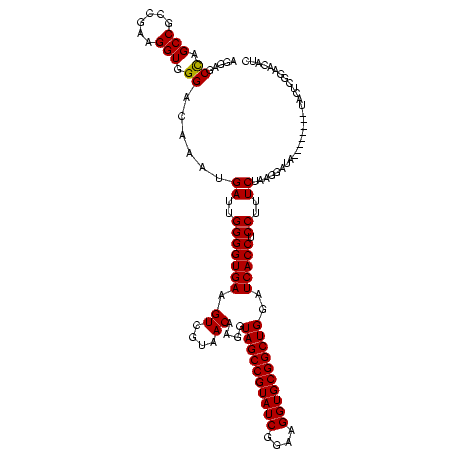
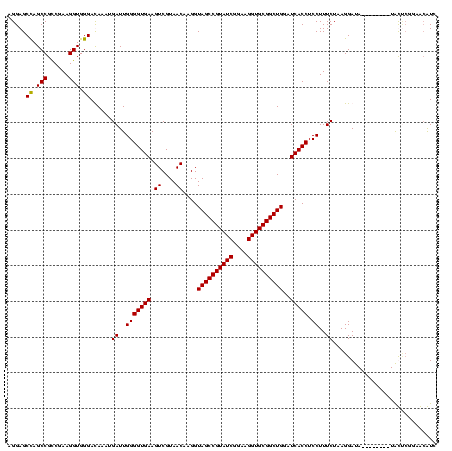
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -42.88 |
| Energy contribution | -41.88 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/160-280 CGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACC-GUAAGGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((-....)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... ( -46.00) >sp_ag.0 1908203 120 - 2160267/160-280 CGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((...((.((((..(((..((.....)).))).)))).))..))))..))))))..)))...................((((((((((....))))))))))... ( -43.50) >sy_ha.0 2542316 119 + 2685015/160-280 CGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUU-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..(((((((...((((..(((..((....-)).))).)))))))..))))..))))))..)))...................((((((((((....))))))))))... ( -43.70) >sy_au.0 2278565 120 - 2809422/160-280 CGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCUUUUAGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..(((((((...((((..(((..((.....)).))).)))))))..))))..))))))..)))...................((((((((((....))))))))))... ( -43.60) >consensus CGAGAGUUUGUAACACCCGAAGCCGGUGAAGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU (((..((((((..((((....(.((((..(((..((.....)).))).)))).)...))))..))))))..)))...................((((((((((....))))))))))... (-42.88 = -41.88 + -1.00) # Strand winner: reverse (1.00)

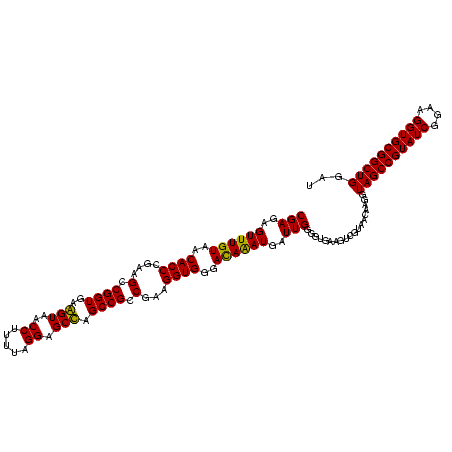
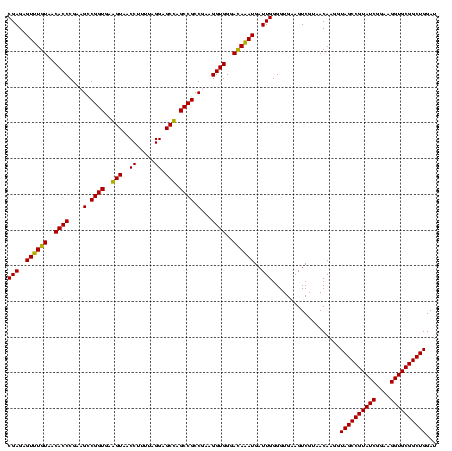
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.28 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -38.95 |
| Energy contribution | -35.58 |
| Covariance contribution | -3.38 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.64 |
| Structure conservation index | 1.06 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/360-480 UGACCUGGGCUACACACGUGCUACAAUGGCUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAGUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAA .((.((((((.((....))))).....(((((((.....(((..((..((((....))))..))....))).....))))))).))).))....(((((((........))))))).... ( -42.20) >sp_ag.0 1908203 120 - 2160267/360-480 UGACCUGGGCUACACACGUGCUACAAUGGUUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAAUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAA .((.((((((.((....))))).....(((((((.....(((..((..((((....))))..))....))).....))))))).))).))....(((((((........))))))).... ( -39.90) >sy_ha.0 2542316 120 + 2685015/360-480 UGAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAA .((((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..))))))))))((((((........))))))..... ( -32.50) >sy_au.0 2278565 120 - 2809422/360-480 UGAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAA .((((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..))))))))))((((((........))))))..... ( -32.50) >consensus UGACCUGGGCUACACACGUGCUACAAUGGACAAUACAAAGAGCAGCAAAACCGCGACGGCAAGCAAAUCCCAUAAAGCCAUUCUCAGUUCGGAUUGUAGGCUGCAACUCGACUACAUGAA .((((((((((.((....)).......(((((((.....(((..((..((((....))))..))....))).....)))))))..))))))))))((((((........))))))..... (-38.95 = -35.58 + -3.38) # Strand winner: reverse (0.90)

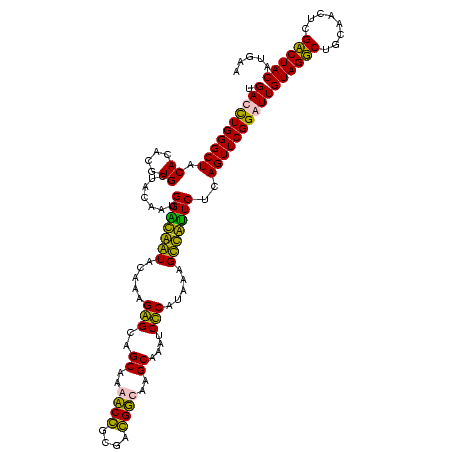
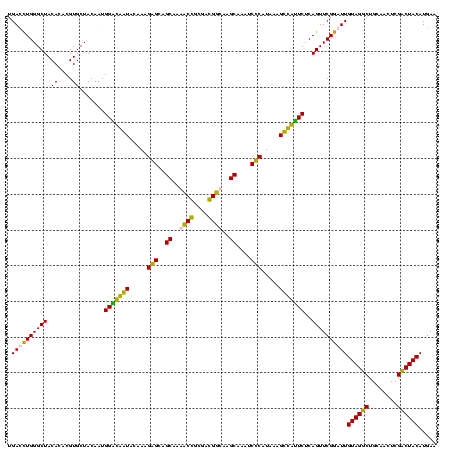
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.03 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -41.67 |
| Energy contribution | -44.42 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/480-600 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUGAAUGAUGGCAACUAACAAUAGGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......))))))))))........(((((.....))))).((((((((..((((((.(..(((......(....)......)))..)..))))))...)))))))) ( -47.70) >sp_ag.0 1908203 120 - 2160267/480-600 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAGGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......))))))))))........(((((.....))))).((((((((..((((((....((((.((..(....)...)).))))....))))))...)))))))) ( -49.50) >sy_ha.0 2542316 120 + 2685015/480-600 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -40.90) >sy_au.0 2278565 120 - 2809422/480-600 UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......)))))))))).((((.(((((((.....))))).))....(((((..((((((......))..)))).)))))...))))(((.(((.....))).))). ( -40.90) >consensus UAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAAGGGUUGCGCUCGUUGCGGGACU ...((((.((((((......))))))))))........(((((.....))))).((((((((..((((((....(((((((..(....)...)))))))....))))))...)))))))) (-41.67 = -44.42 + 2.75)

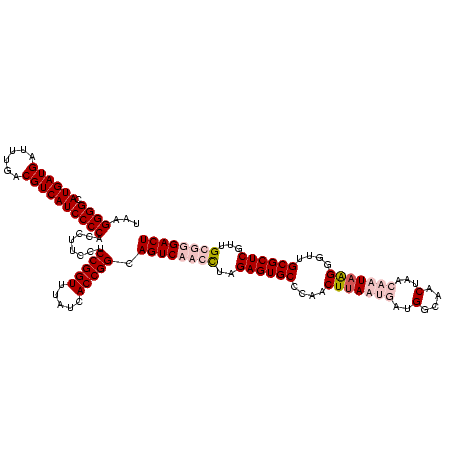
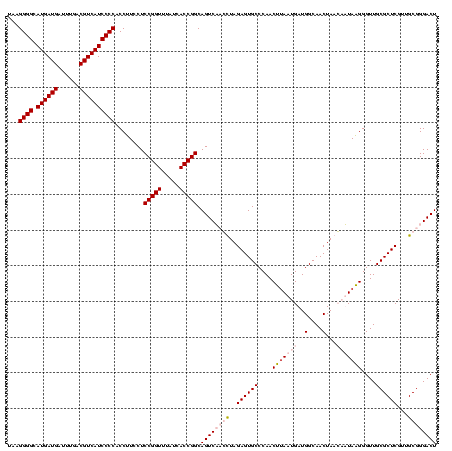
| Location | 1,854,960 – 1,855,077 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.66 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -36.20 |
| Energy contribution | -35.70 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694553 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 117 + 2038615/600-720 GAACCUUACCAGGUCUUGACAUCCCUCUGACCGCUCUAGAGAUAGAG--UUUUCCUUCGGGACA-GAGGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ..((((....))))((..(((((((((((...((((((....)))))--)..(((....)))))-)))).((((.(.(.((.(((.....))))).).).))))....)))))..))... ( -42.50) >sp_ag.0 1908203 117 - 2160267/600-720 GAACCUUACCAGGUCUUGACAUCCUUCUGACCGGCCUAGAGAUAGGC--UUUCUCUUCGGAGCA-GAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA ..((((....))))((..(((((((((((...((((((....)))))--)..(((....)))))-)))).((((.(.(.((.(((.....))))).).).))))....)))))..))... ( -40.70) >sy_ha.0 2542316 119 + 2685015/600-720 GAACCUUACCAAAUCUUGACAUCCUU-UGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA .((((.(((((...((((...(((((-((....(((((....)))))...(((((....)))))))))).)))))))))))...))))......((((((..((....))..).))))). ( -40.10) >sy_au.0 2278565 119 - 2809422/600-720 GAACCUUACCAAAUCUUGACAUCCUU-UGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA .((((.(((((...((((...(((((-((....(((((....)))))...(((((....)))))))))).)))))))))))...))))......((((((..((....))..).))))). ( -40.10) >consensus GAACCUUACCAAAUCUUGACAUCCUU_UGACAACUCUAGAGAUAGAG__UUCCCCUUCGGGGCA_AAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUA .(((((...((..((.((((((....(((((..(((((....)))))...(((((....)))))......(((((.((.....)).))))))))))...)))))).))..))..))))). (-36.20 = -35.70 + -0.50) # Strand winner: reverse (1.00)

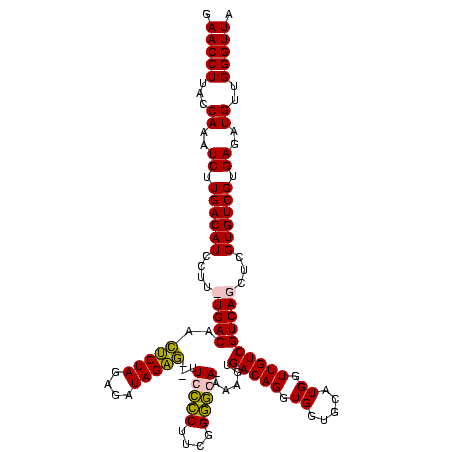
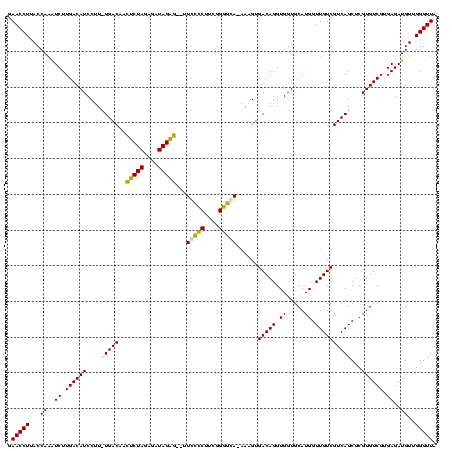
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -54.25 |
| Consensus MFE | -56.62 |
| Energy contribution | -54.25 |
| Covariance contribution | -2.38 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.85 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/760-880 CGAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCA (((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))).. ( -53.90) >sp_ag.0 1908203 120 - 2160267/760-880 CGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCA (((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))).. ( -55.90) >sy_ha.0 2542316 120 + 2685015/760-880 CGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCA (((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))).. ( -53.60) >sy_au.0 2278565 120 - 2809422/760-880 CGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCA (((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))).. ( -53.60) >consensus CGAUGAGUGCUAAGUGUUAGGCCCUUUCCGCCCCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCA (((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..))).)))).(((....))).. (-56.62 = -54.25 + -2.38) # Strand winner: reverse (0.99)

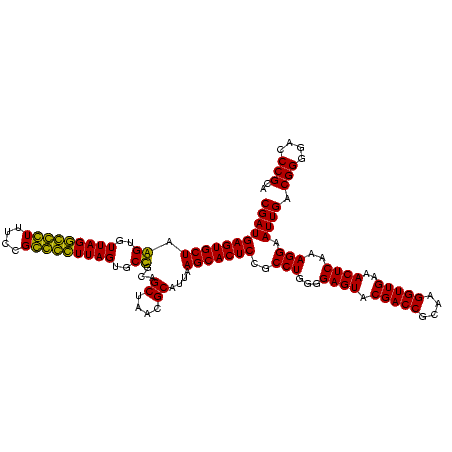
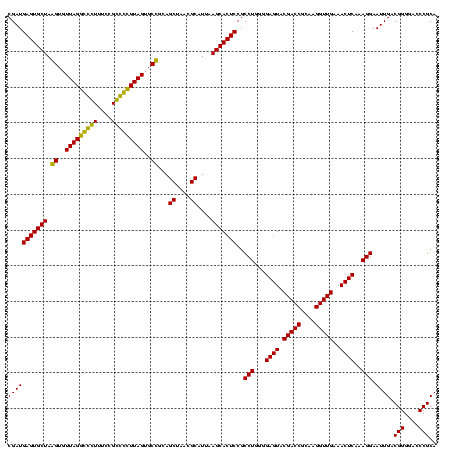
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -45.75 |
| Consensus MFE | -48.31 |
| Energy contribution | -45.75 |
| Covariance contribution | -2.56 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.56 |
| Structure conservation index | 1.06 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/800-920 AGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCUGUAAACGAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGA ......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).. ( -44.20) >sp_ag.0 1908203 120 - 2160267/800-920 AGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGA ......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).. ( -47.80) >sy_ha.0 2542316 120 + 2685015/800-920 AUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGA ......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).. ( -45.50) >sy_au.0 2278565 120 - 2809422/800-920 AUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGA ......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).. ( -45.50) >consensus AGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGCCCUUUCCGCCCCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGA ......((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).. (-48.31 = -45.75 + -2.56) # Strand winner: reverse (0.98)

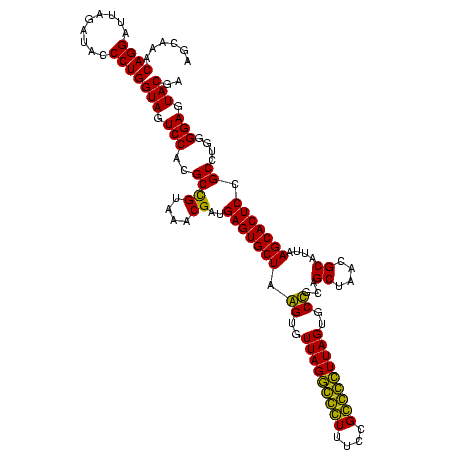
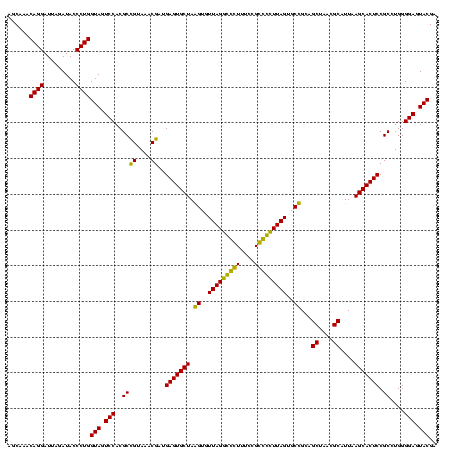
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -45.10 |
| Consensus MFE | -45.55 |
| Energy contribution | -44.30 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.87 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/920-1040 UGCAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGCUUGUAACUGACGCUGAGGCUCGAAAGCGUGGGG ((((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..)))))).((.(((((..........))))).)). ( -46.60) >sp_ag.0 1908203 120 - 2160267/920-1040 UGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCGAAAGCGUGGGG ((((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..)))))).((.(((((..........))))).)). ( -47.40) >sy_ha.0 2542316 120 + 2685015/920-1040 UGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGG ((((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..)))))).((.(((((..........))))).)). ( -43.20) >sy_au.0 2278565 120 - 2809422/920-1040 UGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGG ((((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..)))))).((.(((((..........))))).)). ( -43.20) >consensus UGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAAGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGG ((((((...((((((((...((((((((((.(((........)))..))))))))))...)))..((.((....)).)))))))..)))))).((.(((((..........))))).)). (-45.55 = -44.30 + -1.25) # Strand winner: reverse (1.00)

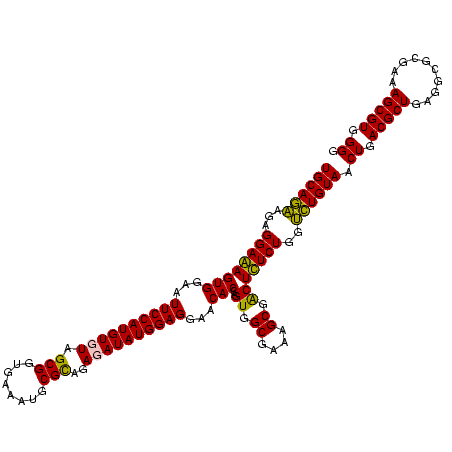
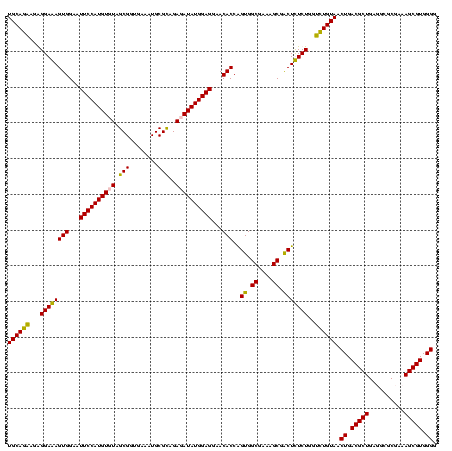
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.87 |
| Mean single sequence MFE | -34.43 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.65 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/960-1080 GCUUAACCAUAGUAGG-CUUUGGAAACUGUUUAACUUGAGUGCAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCU ((((..((....((((-(...(....).))))).((((....))))..))..))))((..((((((((((.(((......)))....)))))))))).....))(((.((....)).))) ( -35.60) >sp_ag.0 1908203 119 - 2160267/960-1080 GCUUAACCAUUGUACG-CUUUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCU ((((..((.((((((.-(((((....)..))))......))))))...))..))))((..((((((((((.(((......)))....)))))))))).....))(((.((....)).))) ( -36.30) >sy_ha.0 2542316 120 + 2685015/960-1080 GCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACU ((((((((.....)).((...(....)..))....)))))).............(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).))) ( -32.90) >sy_au.0 2278565 120 - 2809422/960-1080 GCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACU ((((((((.....)).((...(....)..))....)))))).............(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).))) ( -32.90) >consensus GCUCAACCAUGGAAGG_CAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAAGCGACU ((((((((......)).....(....)........)))))).............(((...((((((((((.(((........)))..))))))))))...))).(((.((....)).))) (-28.90 = -28.65 + -0.25) # Strand winner: reverse (1.00)

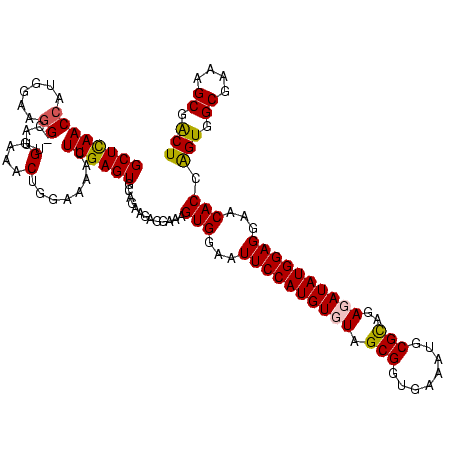
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.54 |
| Mean single sequence MFE | -34.72 |
| Consensus MFE | -35.59 |
| Energy contribution | -32.40 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/1000-1120 GAGCGCAGGCGGUUAGAUAAGUCUGAAGUUAAAGGCUGUGGCUUAACCAUAGUAGG-CUUUGGAAACUGUUUAACUUGAGUGCAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAA ..(((((..((((((((((..((..(((((....(((((((.....))))))).))-)))..))...)))))))).))..)))...........((((....)))).....))....... ( -35.90) >sp_ag.0 1908203 119 - 2160267/1000-1120 GAGCGCAGGCGGUUCUUUAAGUCUGAAGUUAAAGGCAGUGGCUUAACCAUUGUACG-CUUUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAA ..(((((..((((((((((..((..((((.....(((((((.....)))))))..)-)))..))...)))))))).))..)))...........((((....)))).....))....... ( -35.80) >sy_ha.0 2542316 120 + 2685015/1000-1120 GCGCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAA .((((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....)))...... ( -33.60) >sy_au.0 2278565 120 - 2809422/1000-1120 GCGCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAA .((((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....)))...... ( -33.60) >consensus GAGCGCAGGCGGUUUUUUAAGUCUGAAGUGAAAGCCCACGGCUCAACCAUGGAAGG_CAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAA ..(((((..((((((((((..((..(((......(((((((.....)))))))....)))..))...)))))))).))..)))...........((((....)))).....))....... (-35.59 = -32.40 + -3.19) # Strand winner: reverse (1.00)

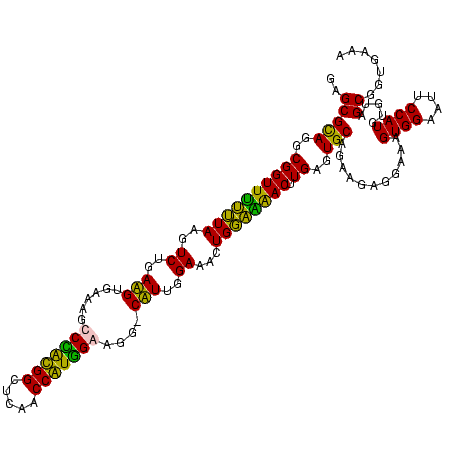
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -33.72 |
| Energy contribution | -30.73 |
| Covariance contribution | -3.00 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/1200-1320 ACGGAAGUCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUAAGAGAAGAACGAGUGUGAGAGUGGAAAGUUCACACUGUGACGGUAUCUUACCAG ..(((((....((((.(((...((((....))(((((...))))))).((((....((((......))))....))))(((((((..(....).))))))))))..))))..))).)).. ( -32.70) >sp_ag.0 1908203 120 - 2160267/1200-1320 ACGGAAGUCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGAGAAGAACGUUGGUAGGAGUGGAAAAUCUACCAAGUGACGGUAACUAACCAG ..((.(((...((((((((((..(((....)))....(((((((((...)).))))).))))))))........((.((((((((..(....).))))))))))..)))).)))..)).. ( -30.10) >sy_ha.0 2542316 119 + 2685015/1200-1320 GCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGU-AACUAUGCACGUCUUGACGGUACCUAAUCAG ((....)).((((((((...((.(((....))).))..((((....))))......))))))))(((((......((.(((((((...-.....)))))))..))......))))).... ( -37.40) >sy_au.0 2278565 119 - 2809422/1200-1320 GCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUAUGUGUAAGU-AACUGUGCACAUCUUGACGGUACCUAAUCAG ((....)).((((((((...((.(((....))).))..((((....))))......))))))))(((((......((.(((((((...-.....)))))))..))......))))).... ( -36.00) >consensus ACGAAAGCCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGAGAAGAACAUAUGUGUAAGU_AAAAAUGCACAUCGUGACGGUACCUAACCAG ((....))..((((((((.....(((....))).....((((....)))).....))))))))((((((......((((((((((.........)))))))).))......))))))... (-33.72 = -30.73 + -3.00) # Strand winner: reverse (1.00)

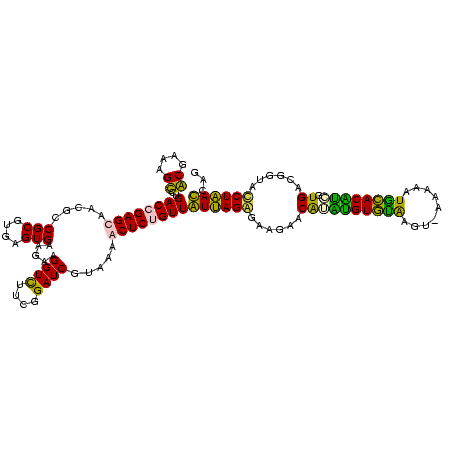
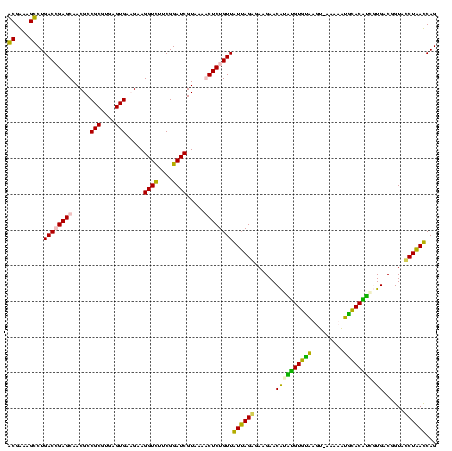
| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -43.68 |
| Consensus MFE | -37.95 |
| Energy contribution | -38.70 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984576 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/1240-1360 ACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCGGCAAUGGACGGAAGUCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUAAGAGAAGAACGAGU .((((....)))).........(..((((((((((...((((....))))(......)...)))((....))...)))))))..)...((((....((((......))))....)))).. ( -41.50) >sp_ag.0 1908203 120 - 2160267/1240-1360 ACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCGGCAAUGGACGGAAGUCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGAGAAGAACGUUG .((((....)))).((((....(..((((((((((...((((....))))(......)...)))((....))...)))))))..).....((....(((((....)))))....)))))) ( -41.40) >sy_ha.0 2542316 120 + 2685015/1240-1360 ACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUAC .((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))...... ( -45.90) >sy_au.0 2278565 120 - 2809422/1240-1360 ACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUAU .((((....))))..............((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....))))))...... ( -45.90) >consensus ACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGCCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGAGAAGAACAUAU .((((....))))..............((((((.....((((....))))((((((((.....(((....))).....((((....)))).....)))))))).....))))))...... (-37.95 = -38.70 + 0.75) # Strand winner: reverse (1.00)

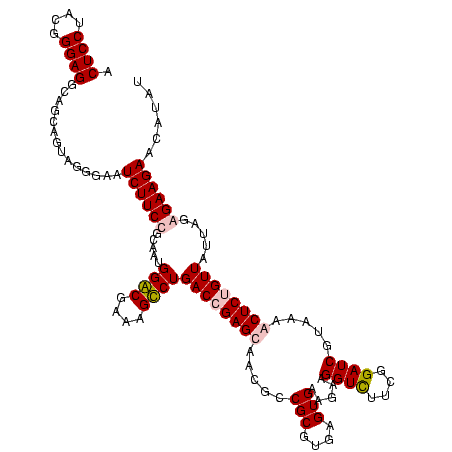

| Location | 1,854,960 – 1,855,080 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -38.23 |
| Energy contribution | -37.16 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 120 + 2038615/1360-1480 CUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGUGA (((((.(........).))))).((((((((((.........))))))))))...(((..(((..((((..(((((........)))..((.........)).))..)))).)))..))) ( -37.10) >sp_ag.0 1908203 120 - 2160267/1360-1480 CUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGA .((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).))))))........... ( -39.80) >sy_ha.0 2542316 120 + 2685015/1360-1480 CUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAACUAAUACGGCGCGGGUCCAUCUAUAAGUGA .(((((((((((...........((((((((((.........))))))))))((((.......((((((((.......)))))))).......))))))))).))))))........... ( -43.34) >sy_au.0 2278565 120 - 2809422/1360-1480 CUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGA .((((((((((..((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...))..)))))).))))........... ( -42.40) >consensus CUGGACCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCCGUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGUGA .((((((((((............((((((((((.........))))))))))((((.......((((((((.......)))))))).......)))).)))).))))))........... (-38.23 = -37.16 + -1.06)

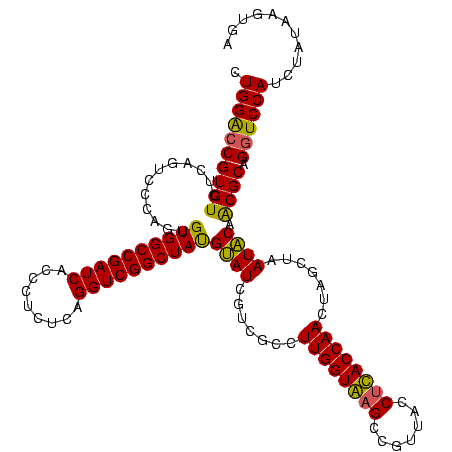
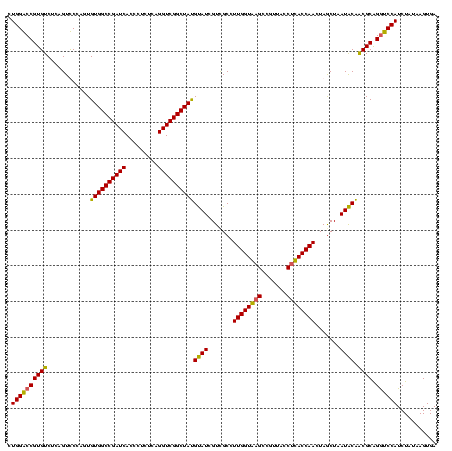
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.91 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -32.54 |
| Energy contribution | -28.23 |
| Covariance contribution | -4.31 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825676 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/1400-1520 UGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGCA-UCACUACCAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUACAUAGCCGACCUG .(((((((....))))))).......(((((((..(.((-((.......)))).)..)))..((((((.(((..((((((((.......)))))))))))...))))))......)))). ( -32.70) >sp_ag.0 1908203 119 - 2160267/1400-1520 UAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU-UCACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGAGGUAAAGGCUCACCAAGGCGACGAUACAUAGCCGACCUG ((((((((....)))))))).......(((((....)))-)).........(((...((..((((....(((..((((((((.......))))))))))).))))..))...)))..... ( -33.00) >sy_ha.0 2542316 120 + 2685015/1400-1520 UUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGUUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGACCUG ...(((((....)))))(((((..(((.....)))..)))))..........((..(.((..((((((.(((..((((((((.......))))))))...)))))))))..)).).)).. ( -34.60) >sy_au.0 2278565 120 - 2809422/1400-1520 UUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCAUAGCCGACCUG ((((((((....)))))))).((((..(((((.....)))))..))))....((.((.((..((((((.(((..((((((((.......)))))))))))...))))))..)).)))).. ( -38.70) >consensus UUAGAACCGCAUGGUACUAAAGUAAAAGAUGCACUUGCU_UCACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUAAGGUAACGGCUCACCAAGGCGACGAUACAUAGCCGACCUG .(((((((....)))))))........(((((.....)))))..........((....((..((((((.(((..((((((((.......)))))))))))...))))))..))...)).. (-32.54 = -28.23 + -4.31) # Strand winner: reverse (1.00)

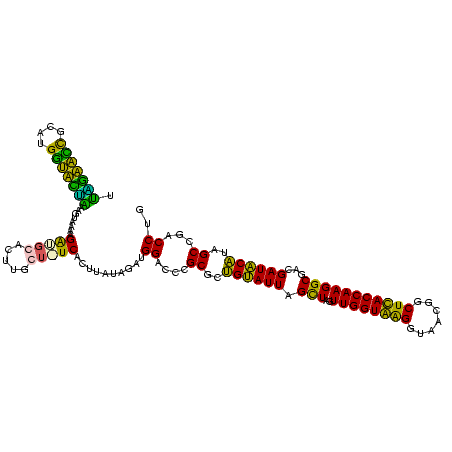
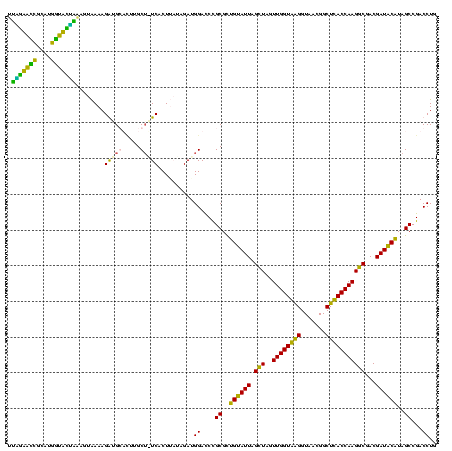
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -18.56 |
| Energy contribution | -17.75 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928240 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/1440-1560 CACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGUGA-UGCAAGUGCACCUUUUAAGCAAAUGUCAUGCAACAUCCACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUCCC .......((((((((((..(((((((.(((...(((....-)))..))).)))......(((.......)))..............)))))))))))))).................... ( -26.70) >sp_ag.0 1908203 119 - 2160267/1440-1560 CACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGA-AGCAAUUGCUCCUUUUAAAUAACUAACAUGUGUUAAUUACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUCCC .......((((((((((..((((((......))..(((((-(((....))).........((((........)))).)))))....)))))))))))))).................... ( -20.80) >sy_ha.0 2542316 120 + 2685015/1440-1560 UACCAACUAACUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCC ....((((..(((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......))))))))))))...(((....))).))))..... ( -28.20) >sy_au.0 2278565 120 - 2809422/1440-1560 UACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCC ....(((((((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....)))))))))))))).(((....))).))))..... ( -33.00) >consensus CACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGUGA_AGCAAGACCACCUUUCAAUUAAGAACCAUGCGGUACAAAAUAUUAUCCGGUAUUAGCUACCGUUUCCAAAAGUUAUCCC ....(((((((((((((...((...(((........).))..))...............(((((((((....))))))))).........))))))))).(((....))).))))..... (-18.56 = -17.75 + -0.81)


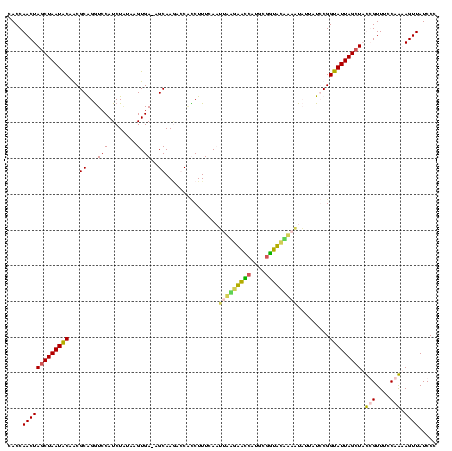
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.55 |
| Mean single sequence MFE | -38.38 |
| Consensus MFE | -32.88 |
| Energy contribution | -28.62 |
| Covariance contribution | -4.25 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.21 |
| SVM RNA-class probability | 0.999838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/1440-1560 GGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGCA-UCACUACCAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUG ....(((((((((....))))(((((((((((((...(((..((((((....))))))..)))....(((((....)))-))....((....))...))))..))))))))))))))... ( -40.20) >sp_ag.0 1908203 119 - 2160267/1440-1560 GGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU-UCACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUG ....(((((((((....))))(((((((((((((...(((((((((((....)))))))))))....(((((....)))-))...............))))..))))))))))))))... ( -37.10) >sy_ha.0 2542316 120 + 2685015/1440-1560 GGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGUUAGUUGGUA (((....((((((....))))))......(((.((((......(((((....)))))(((((..(((.....)))..)))))..))))...))).)))..((((...........)))). ( -37.60) >sy_au.0 2278565 120 - 2809422/1440-1560 GGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUA .......((((((....))))))....((((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))..(((((....)).))).)))) ( -38.60) >consensus GGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAGAGUUAGAACCGCAUGGUACUAAAGUAAAAGAUGCACUUGCU_UCACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUA ....(((((((((....))))((((((((((((......(((((((((....)))))))))................(((((.......)))))....)))..))))))))))))))... (-32.88 = -28.62 + -4.25) # Strand winner: reverse (1.00)

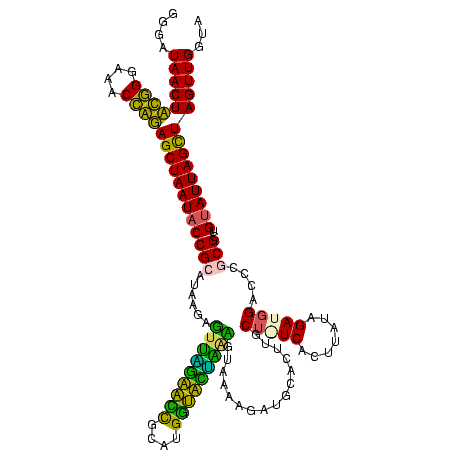
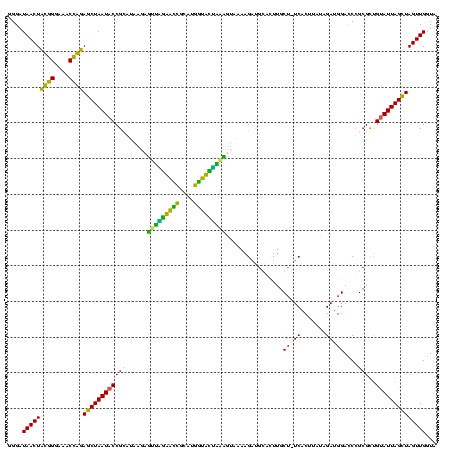
| Location | 1,854,960 – 1,855,079 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.71 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -33.38 |
| Energy contribution | -27.75 |
| Covariance contribution | -5.62 |
| Combinations/Pair | 1.76 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.03 |
| SVM RNA-class probability | 0.999767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 119 + 2038615/1480-1600 GAACGGGUGAGUAACGCGUAGGUAACCUGCCUGGUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGCA- ......(..(((........((((.(((((......)))))......((((((....))))))....))))((((..(((..((((((....))))))..))).....)))))))..).- ( -41.30) >sp_ag.0 1908203 119 - 2160267/1480-1600 GAACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU- ..(((.((.....)).))).(((((..(((..(...((((....((.((((((....)))))).))...))))....(((((((((((....)))))))))))....)..))).)))))- ( -36.70) >sy_ha.0 2542316 120 + 2685015/1480-1600 GGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUA ......(((.....)))(((((((...)))))))....((((..((.((((((....)))))).))...))))........(((((((....)))))))(((..(((.....)))..))) ( -37.10) >sy_au.0 2278565 120 - 2809422/1480-1600 GGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUG (((((.((.....)).)))((....))..))..((((((((((.((.((((((....)))))).))...))........(((((((((....)))))))))........))))))))... ( -35.80) >consensus GAACGGGUGAGUAACACGUAGGUAACCUACCUAUAAGACGGGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAGAGUUAGAACCGCAUGGUACUAAAGUAAAAGAUGCACUUGCU_ .....((..((......((((.....))))........((((..((.((((((....)))))).))...))))......(((((((((....)))))))))............))..)). (-33.38 = -27.75 + -5.62) # Strand winner: reverse (1.00)

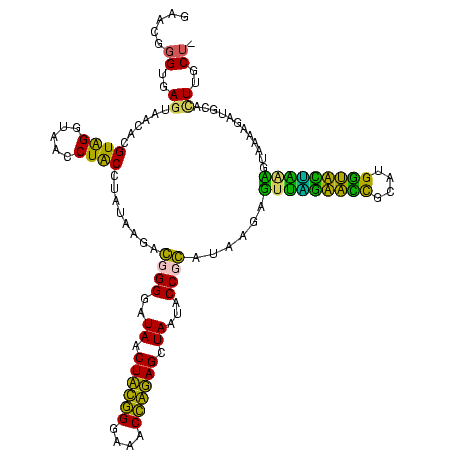
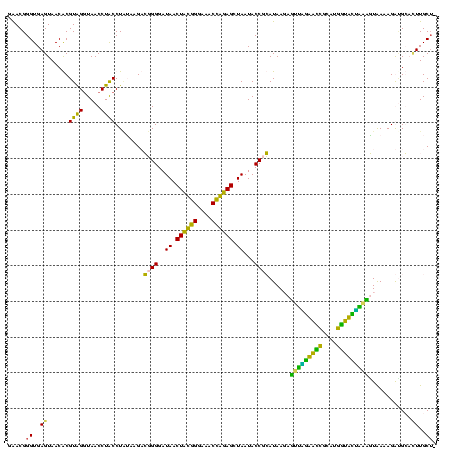
| Location | 1,854,960 – 1,855,078 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -28.20 |
| Energy contribution | -24.64 |
| Covariance contribution | -3.56 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 118 + 2038615/1520-1640 AACGCUGAAG-GAGGAGCUUGC-UUCUCUGGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUGGUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAG .((((....(-((((((....)-))))))......(((((..(....)..))))))))).((((.(((((......)))))......((((((....))))))....))))......... ( -38.80) >sp_ag.0 1908203 120 - 2160267/1520-1640 AACGCUGAGGUUUGGUGUUUACACUAGACUGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAG .((((...(((((((((....))))))))).....(((((..(....)..))))))))).((((.(((((......)))))......((((((....))))))....))))......... ( -40.40) >sy_ha.0 2542316 120 + 2685015/1520-1640 AGCGAACAGACAAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAU .............(..(((..(((.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))....... ( -37.30) >sy_au.0 2278565 120 - 2809422/1520-1640 AGCGAACGGACGAGAAGCUUGCUUCUCUGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAU ......(((..((((((....))))))...(((((((((....((.(((.....))).)).....))..((((......)))).....(((((....)))))))))))))))........ ( -36.80) >consensus AACGAACAGACGAGGAGCUUGCUCCUUUGAGAUGAGCGGCGAACGGGUGAGUAACACGUAGGUAACCUACCUAUAAGACGGGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAGAG .((........((((((....))))))........)).....(((.((.....)).)))(((((...)))))......((((..((.((((((....)))))).))...))))....... (-28.20 = -24.64 + -3.56) # Strand winner: reverse (1.00)

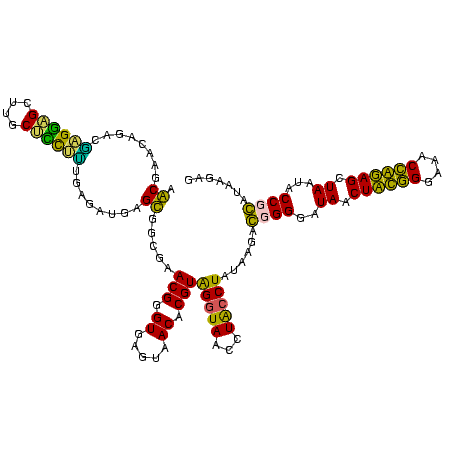
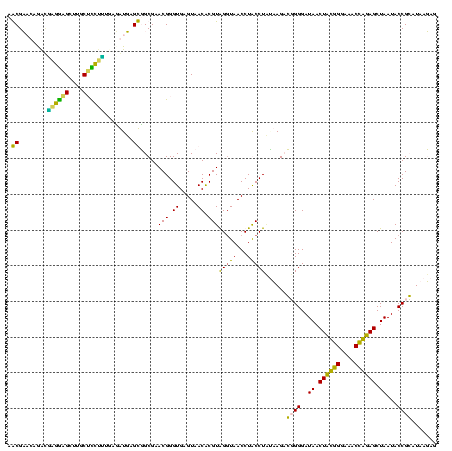
| Location | 1,854,960 – 1,855,074 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.61 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -18.86 |
| Energy contribution | -18.43 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1854960 114 + 2038615/1640-1760 G--AAAAAAACAAUAAAUCUGUCAGU--GACAGAAAUGAGUGAGAACUCA--AACUUUUAAUGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAG .--................(((((((--(.......(((((.((...(((--(((((((...))))))))))..)).))))).......))))))))(((((.......)))))...... ( -32.84) >sp_ag.0 1908203 117 - 2160267/1640-1760 GACAAAAAAACAAUAAAUCUGUCAGCGAGACAGAAAAGAGC-AAAGCUCA--AACUUUUAAUGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAG .................((((((.....))))))...((((-...(.(((--(((((((...)))))))))).)...))))(((....((((.....)))).)))..(((......))). ( -31.60) >sy_ha.0 2542316 115 + 2685015/1640-1760 A--AAUGUUACAA-ACACUAUUUAGUA--UUAUGAGCUAAUCAAACAUCAUAAAUUUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCG .--..........-..........(((--(((((((((((((((((.((((((....))))))...)))))))..)))))))((....((((.....)))).)))))))).......... ( -29.10) >sy_au.0 2278565 115 - 2809422/1640-1760 A--UAAGUUACAA-ACAUUAUUUAGUA-UUUAUGAGCUAAUCAAACAUCAU-AAUUUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCG .--..........-..........(((-(.((((((((((((((((.((((-(.....)))))...)))))))..))))))(((....((((.....)))).))).))).))))...... ( -27.20) >consensus A__AAAAAAACAA_AAAUCAGUCAGUA_GACAGAAACGAAUCAAACAUCA__AACUUUUAAGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAG ....................((((((............................(((.....))).(((((.(((((...)))))))))))))))).(((((.......)))))...... (-18.86 = -18.43 + -0.44) # Strand winner: reverse (1.00)

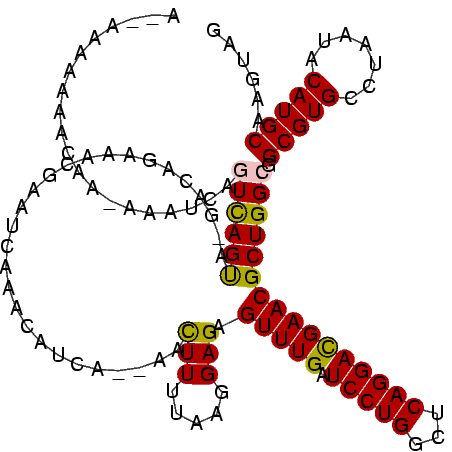
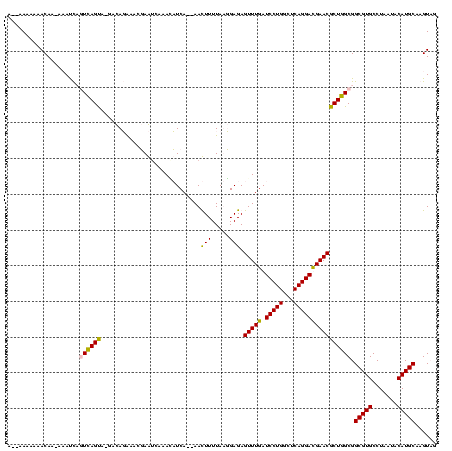
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:01:07 2006