
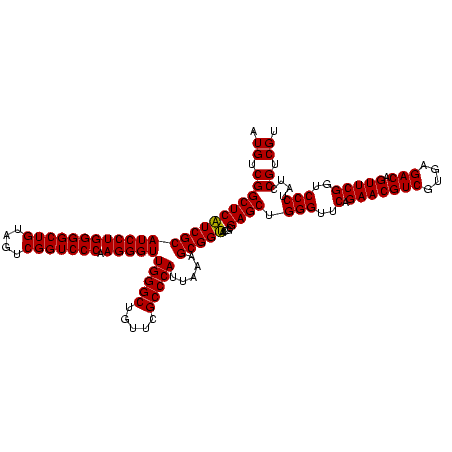
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 2,539,203 – 2,539,323 |
| Length | 120 |
| Max. P | 0.999893 |

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -53.86 |
| Consensus MFE | -53.88 |
| Energy contribution | -53.40 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.24 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/280-400 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGU (((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).))) ( -53.30) >sy_au.0 2275427 120 - 2809422/280-400 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGU (((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).))) ( -53.30) >sy_sa.0 1670096 120 - 2516575/280-400 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGU (((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).))) ( -53.30) >sp_pn.0 1851953 120 + 2038615/280-400 AUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGC ..(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....)))))).(((..(.(((((((....))).)))))..)))....)).).. ( -54.70) >sp_ag.0 1905165 120 - 2160267/280-400 AUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGC ..(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....)))))).(((..(.(((((((....))).)))))..)))....)).).. ( -54.70) >consensus AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGU .((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).)). (-53.88 = -53.40 + -0.48) # Strand winner: reverse (0.99)

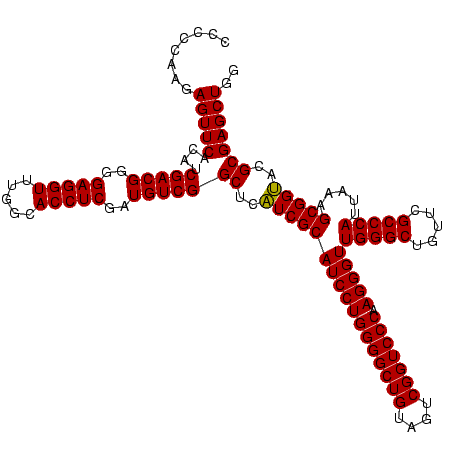
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -53.70 |
| Consensus MFE | -54.18 |
| Energy contribution | -53.70 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/320-440 CCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGG ........(((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..))))))).. ( -53.10) >sy_au.0 2275427 120 - 2809422/320-440 CCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGG ........(((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..))))))).. ( -53.10) >sy_sa.0 1670096 120 - 2516575/320-440 CCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGG ........(((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..))))))).. ( -53.10) >sp_pn.0 1851953 120 + 2038615/320-440 CCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGG ........(((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..))))))).. ( -54.60) >sp_ag.0 1905165 120 - 2160267/320-440 CCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGG ........(((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..))))))).. ( -54.60) >consensus CCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGG ........(((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..))))))).. (-54.18 = -53.70 + -0.48) # Strand winner: reverse (0.99)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -44.82 |
| Consensus MFE | -45.30 |
| Energy contribution | -44.82 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.63 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/360-480 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >sy_au.0 2275427 120 - 2809422/360-480 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >sy_sa.0 1670096 120 - 2516575/360-480 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >sp_pn.0 1851953 120 + 2038615/360-480 UCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -43.50) >sp_ag.0 1905165 120 - 2160267/360-480 UCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -43.50) >consensus UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... (-45.30 = -44.82 + -0.48) # Strand winner: forward (0.61)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -37.72 |
| Consensus MFE | -37.66 |
| Energy contribution | -35.82 |
| Covariance contribution | -1.84 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.10 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/480-600 GGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGC ....(((......((((.((((.(..((((........))))..((((..(((......)))..)))).(((....)))).)))).)))))))((((((.((........)))))))).. ( -36.80) >sy_au.0 2275427 120 - 2809422/480-600 GGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGC ....(((......((((.((((.(..((((........))))..((((..(((......)))..)))).(((....)))).)))).)))))))((((((.((........)))))))).. ( -36.80) >sy_sa.0 1670096 120 - 2516575/480-600 GGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGC ....(((......((((.((((.(..((((........))))..((((..((((....))))..)))).(((....)))).)))).)))))))((((((.((........)))))))).. ( -38.00) >sp_pn.0 1851953 120 + 2038615/480-600 GGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGC ....(((......((((.(((((((..(((.((((....)))))))((....))))).))))((((...(((....)))..)))).)))))))((((((.((........)))))))).. ( -39.30) >sp_ag.0 1905165 120 - 2160267/480-600 GGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGC ....(((..(((......(((((((..(((.((((....)))))))((....))))).))))((((...(((....)))..))))..))))))((((((.((........)))))))).. ( -37.70) >consensus GGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGC ....(((......((((.((((.(..((((........))))..((((..(((......)))..)))).(((....)))).)))).)))))))((((((.((........)))))))).. (-37.66 = -35.82 + -1.84) # Strand winner: reverse (1.00)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -36.54 |
| Energy contribution | -34.30 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.40 |
| SVM RNA-class probability | 0.999889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/520-640 CCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACG (((.....((.......))((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))((((............)))))))))))(((....))). ( -38.00) >sy_au.0 2275427 120 - 2809422/520-640 CCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACG (((.....((.......))((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))((((............)))))))))))(((....))). ( -38.00) >sy_sa.0 1670096 120 - 2516575/520-640 CCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACG ......((((..(...)..))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..((((....))))..)))).(((....))). ( -38.30) >sp_pn.0 1851953 120 + 2038615/520-640 CCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCG (((...........((((.(((....)))..)))).(((((((.....)))))))((.(((((((..(((.((((....)))))))((....))))).))))..)))))(((....))). ( -37.60) >sp_ag.0 1905165 120 - 2160267/520-640 CCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCG (((...........((((.(((....)))..)))).(((((((.....)))))))((.(((((((..(((.((((....)))))))((....))))).))))..)))))(((....))). ( -38.20) >consensus CCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACG .......(((..(...)..)))...(((((((((..(((((((.....))))))).....(((......))).)))))))))..((((..(((......)))..)))).(((....))). (-36.54 = -34.30 + -2.24) # Strand winner: reverse (0.99)

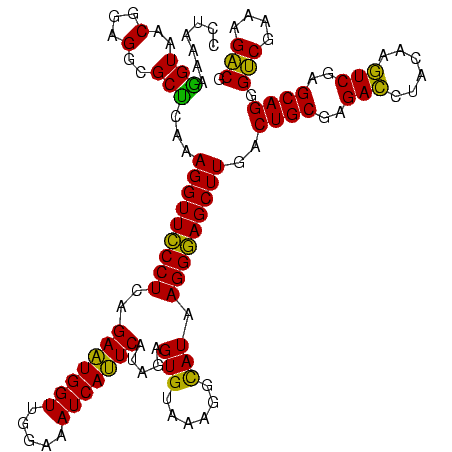
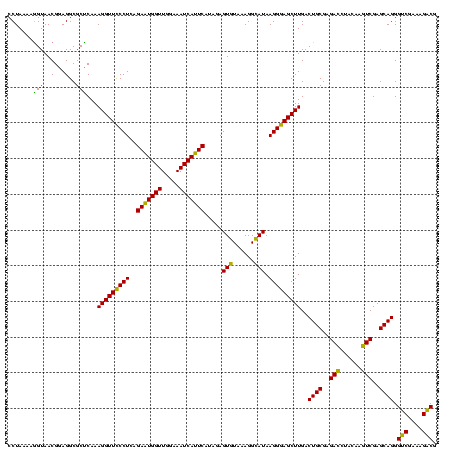
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -26.47 |
| Energy contribution | -26.83 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/560-680 GCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCU (((((((((((......))......(((((.........))))))))))))......(..(((((((...........)))))))..)))....((((..(.....)..))))....... ( -28.50) >sy_au.0 2275427 120 - 2809422/560-680 GCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCU (((((((((((......))......(((((.........))))))))))))......(..(((((((...........)))))))..)))....((((..(.....)..))))....... ( -28.50) >sy_sa.0 1670096 120 - 2516575/560-680 GCUUCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCU (((((((((((......))......(((((.........))))))))))))......(..(((((((...........)))))))..)))....((((...........))))....... ( -25.90) >sp_pn.0 1851953 120 + 2038615/560-680 GCUCCCUUAUACCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCU ..(((((((........(.....).(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))...((((...)))) ( -33.00) >sp_ag.0 1905165 120 - 2160267/560-680 GCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCU ..(((((((................(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))...((((...)))) ( -35.19) >consensus GCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCUGACACUGUCU (((((((((................(((((.........)))))))))))).........(((((((...........)))))))...))....((((.((.....)).))))....... (-26.47 = -26.83 + 0.36)

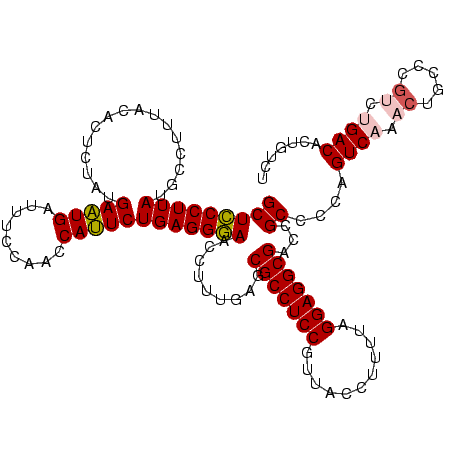
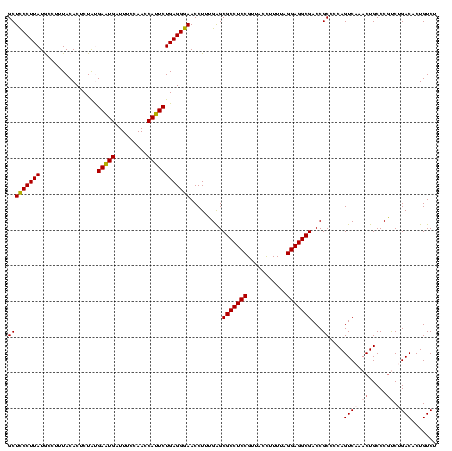
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -40.14 |
| Consensus MFE | -39.78 |
| Energy contribution | -38.66 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.82 |
| SVM RNA-class probability | 0.997230 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/560-680 AGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGC .(((.(((((((((.....)))))))....)).)))((((((...........))))))........(((((((..(((((((.....))))))).....((((....)))).))))))) ( -39.70) >sy_au.0 2275427 120 - 2809422/560-680 AGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGC .(((.(((((((((.....)))))))....)).)))((((((...........))))))........(((((((..(((((((.....))))))).....((((....)))).))))))) ( -39.70) >sy_sa.0 1670096 120 - 2516575/560-680 AGACAGUGUCAGAUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGC .(((...)))...(((((.....(((((...)))))((((((...........)))))))))))...(((((((..(((((((.....))))))).....((((....)))).))))))) ( -37.90) >sp_pn.0 1851953 120 + 2038615/560-680 AGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGC ((((...))))...((((.....(((((...)))))((((((...........))))))))))....(((((((..(((((((.....)))))))((.....)).........))))))) ( -40.70) >sp_ag.0 1905165 120 - 2160267/560-680 AGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGC ((((...))))...((((.....(((((...)))))((((((...........))))))))))....(((((((..(((((((.....)))))))......((((....))))))))))) ( -42.70) >consensus AGACAGUGUCAGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGC .(((...)))....((((.....(((((...)))))((((((...........))))))))))....(((((((..(((((((.....))))))).....(((......))).))))))) (-39.78 = -38.66 + -1.12) # Strand winner: reverse (1.00)

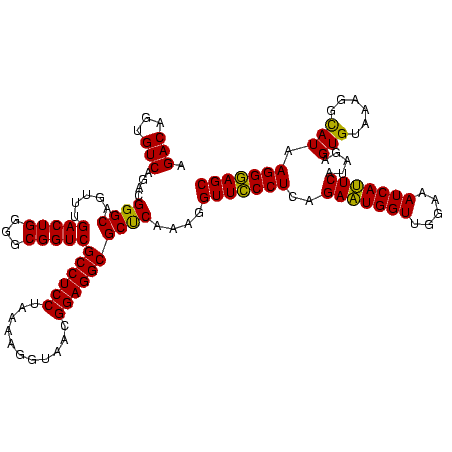
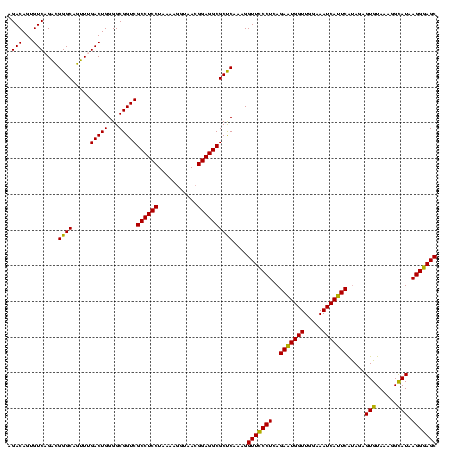
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -40.88 |
| Consensus MFE | -36.94 |
| Energy contribution | -34.58 |
| Covariance contribution | -2.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/600-720 AUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAG ...(((.(((......((..(((((((...........)))))))..)))))))).....((((((((..(((...)))....(((((.....)))))))))).)))............. ( -39.20) >sy_au.0 2275427 120 - 2809422/600-720 AUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAG ...(((.(((......((..(((((((...........)))))))..)))))))).....((((((((..(((...)))....(((((.....)))))))))).)))............. ( -39.20) >sy_sa.0 1670096 120 - 2516575/600-720 AUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAG .......(((......((.((((((((...........))))))(((.......))).....)).))((((((.(((.....((((((.....)))))))))))))))..)))....... ( -35.20) >sp_pn.0 1851953 120 + 2038615/600-720 AUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACA ..((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))((((.((((.((((((.....)))))).)))..).))))......... ( -44.80) >sp_ag.0 1905165 120 - 2160267/600-720 AGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACA .(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((((.....)))))).))).)))............. ( -46.00) >consensus AUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCUGACACUGUCUCCCACCACGAUAACUGGUGCGGGUUAGAAAGCCAACACAG ...(((.(((..........(((((((...........)))))))....)))))).....((((((....(((...)))...((((((.....)))))).))).)))............. (-36.94 = -34.58 + -2.36)

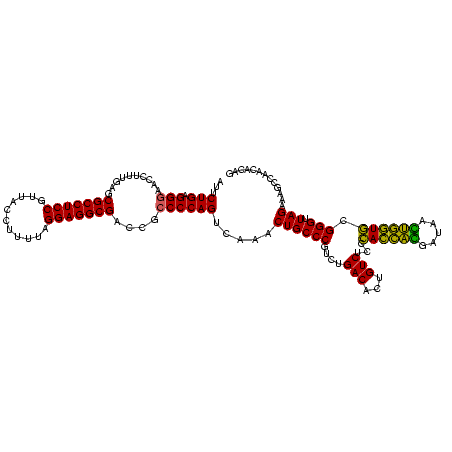
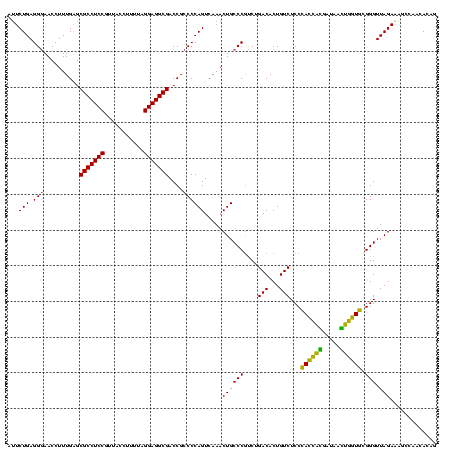
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -34.80 |
| Energy contribution | -29.84 |
| Covariance contribution | -4.96 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.14 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/680-800 CCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGU ((((((((.....)))))).))..............((((((.(((......)))......((((..((((.(((....))).)))).....))))..................)))))) ( -36.40) >sy_au.0 2275427 120 - 2809422/680-800 CCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGU ((((((((.....)))))).))..............((((((.(((......)))......((((..((((.(((....))).)))).....))))..................)))))) ( -36.40) >sy_sa.0 1670096 120 - 2516575/680-800 CCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUCUCAGGCUCCUACCUAUCCUGUACAAGCUGU ((((((((.....)))))).))(((.(....).)))(((((..(((......)))......((((..((((.(((....))).)))).....))))...................))))) ( -34.00) >sp_pn.0 1851953 120 + 2038615/680-800 CCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACACAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGGCGUCCCGAUCUCUUAGACUCCUACCUAUCCUGUACAUGUGGU ((((((((.....)))))).)).........((((((.....(((((((........(((..((......))..)))..(((...........)))......))))))).....)))))) ( -29.90) >sp_ag.0 1905165 120 - 2160267/680-800 CCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGUACAUGUGGU ((((((((.....))))))(((((..((((....(((((....(((......)))....(((((............)))))...))))).....))))..)))))............)). ( -34.00) >consensus CCCACCACGAUAACUGGUGCGGGUUAGAAAGCCAACACAGCAAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUAUCAAAGGCUCCUACCUAUCCUGUACAAGCUGU ((((((((.....)))))).))..............((((((.(((......)))......((((..((((.(((....))).)))).....))))..................)))))) (-34.80 = -29.84 + -4.96)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.17 |
| Mean single sequence MFE | -43.84 |
| Consensus MFE | -38.90 |
| Energy contribution | -33.06 |
| Covariance contribution | -5.84 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/680-800 ACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGG ..((((..(((((..(((...((.(((((....((((((((....)))))))).))))).))(((((....)))))))).)))))..)))).......((.((((((.....)))))))) ( -49.80) >sy_au.0 2275427 120 - 2809422/680-800 ACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGG ..((((..(((((..(((...((.(((((....((((((((....)))))))).))))).))(((((....)))))))).)))))..)))).......((.((((((.....)))))))) ( -49.80) >sy_sa.0 1670096 120 - 2516575/680-800 ACAGCUUGUACAGGAUAGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGG .((((((.(((.......))).))).)))...(((((((((....)))))))))..((((..(((((....)))))..))))..(((((....)))))((.((((((.....)))))))) ( -46.50) >sp_pn.0 1851953 120 + 2038615/680-800 ACCACAUGUACAGGAUAGGUAGGAGUCUAAGAGAUCGGGACGCCAGUUUCGAAGGAGACGCUGUUGGGAUACUACCCUUGUGUUAUGGCCACUCUAACCCAGAUAGGUGAUCCCUAUCGG .((.((((.(((.((..(((((...(((((.((.(((..((....))..))).(....).)).)))))...))))).)).)))))))..............((((((.....)))))))) ( -37.40) >sp_ag.0 1905165 120 - 2160267/680-800 ACCACAUGUACAGGAUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGG .((.........((.(((((((..((((.....((((..((....))..))))..))))......(((......)))...........)))).))).))..((((((.....)))))))) ( -35.70) >consensus ACAGCUUGUACAGGAUAGGUAGGAGCCUAUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCCUGGUGGG ((((((.............(((..((((.....((((((((....))))))))..)))).)))..(((......))).))))))..............((.((((((.....)))))))) (-38.90 = -33.06 + -5.84) # Strand winner: reverse (1.00)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -35.60 |
| Energy contribution | -35.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/840-960 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.((((((((.((((........))))))))((((((((((...(((((.....))))).)))...))))))).........)))))))).)))))............ ( -38.30) >sy_au.0 2275427 120 - 2809422/840-960 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.((((((((.((((........))))))))((((((((((...(((((.....))))).)))...))))))).........)))))))).)))))............ ( -38.30) >sy_sa.0 1670096 120 - 2516575/840-960 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.((((((((.((((........))))))))((((((((((...(((((.....))))).)))...))))))).........)))))))).)))))............ ( -38.30) >sp_pn.0 1851953 120 + 2038615/840-960 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))............ ( -35.60) >sp_ag.0 1905165 120 - 2160267/840-960 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))............ ( -35.60) >consensus UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))............ (-35.60 = -35.60 + 0.00)


| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -43.06 |
| Consensus MFE | -42.40 |
| Energy contribution | -41.44 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/960-1080 GGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC ..(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))........ ( -43.10) >sy_au.0 2275427 120 - 2809422/960-1080 GGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC ..(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))........ ( -43.10) >sy_sa.0 1670096 120 - 2516575/960-1080 GGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC ..(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))........ ( -43.10) >sp_pn.0 1851953 120 + 2038615/960-1080 GGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC (((((..(((..(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))........)))..))))) ( -43.00) >sp_ag.0 1905165 120 - 2160267/960-1080 GGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC (((((..(((..(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))........)))..))))) ( -43.00) >consensus GGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC ((..(..(((..(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))........)))..)..)) (-42.40 = -41.44 + -0.96) # Strand winner: reverse (0.98)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -38.28 |
| Energy contribution | -36.36 |
| Covariance contribution | -1.92 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.49 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989601 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/1000-1120 AAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAAC ............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..))))))))))... ( -39.00) >sy_au.0 2275427 120 - 2809422/1000-1120 AAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAAC ............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..))))))))))... ( -39.00) >sy_sa.0 1670096 120 - 2516575/1000-1120 AAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAAC ............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..))))))))))... ( -39.00) >sp_pn.0 1851953 120 + 2038615/1000-1120 AAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAAC .....(((..((((.(((...(((....)))...))).)))).)))..(((.....)))(((((..((((..((...((((((.((....))..))))))...))..))))))))).... ( -36.10) >sp_ag.0 1905165 120 - 2160267/1000-1120 AAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAAC .....(((..((((.(((...(((....)))...))).)))).)))..(((.....)))(((((..((((..((...((((((.((....))..))))))...))..))))))))).... ( -36.10) >consensus AAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAAC .........(((((((((...(((....)))...))..)))))))...(((.....)))(((((..((((..((...((((((.((....))..))))))...))..))))))))).... (-38.28 = -36.36 + -1.92) # Strand winner: reverse (0.98)


| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.98 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -33.30 |
| Energy contribution | -34.34 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/1120-1240 AGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUC ...((..((((..(((((..((.(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).))).......))..)))))..)))).)) ( -40.40) >sy_au.0 2275427 120 - 2809422/1120-1240 AGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUC ...((..((((..(((((..((.(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).))).......))..)))))..)))).)) ( -39.70) >sy_sa.0 1670096 120 - 2516575/1120-1240 AGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGGAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUC ...((..((((..(((((..((.(((((((......(((((....(....).....)))))..((((..((((....))))..)))))))).))).......))..)))))..)))).)) ( -39.70) >sp_pn.0 1851953 109 + 2038615/1120-1240 AGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCU-----GACUUUAAGUC------AGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUC (((((((....)))))))..........((......(((((....(....).....)))))(((-----(((.....)))------))).)).((((((((((....))..)))))))). ( -36.00) >sp_ag.0 1905165 109 - 2160267/1120-1240 AGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCU-----GAUUUUAGAUC------AGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUC (((((((....)))))))..........((......(((((....(....).....)))))(((-----((((...))))------))).)).((((((((((....))..)))))))). ( -36.50) >consensus AGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAAAGCCCAG_AGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUC ((((((......))))))..........((......(((((....(....).....)))))(((((((.((((....)))).))))))).)).((((((((((....))..)))))))). (-33.30 = -34.34 + 1.04) # Strand winner: reverse (0.98)

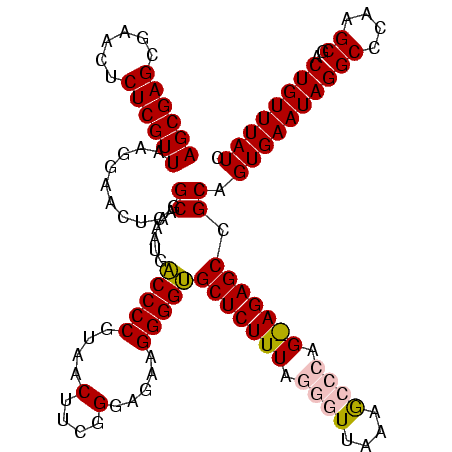
| Location | 2,539,203 – 2,539,321 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.42 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -25.12 |
| Energy contribution | -22.20 |
| Covariance contribution | -2.92 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 118 + 2685015/1360-1480 UUUUAAUCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGAUUGG-AUUGUACGUCUAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGG .((((.((..((.(....).))...)).))))....(((((((((...-)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..))).. ( -31.60) >sy_au.0 2275427 119 - 2809422/1360-1480 UUUUAAUCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUGCGAUUGG-AUUGCACGUCUAAGCAGUAAGGCUGAGUAUUAGGCAAAUCCGGUACUCGUUAAGGCUGAGCUGUGAUGGGG .....(((..((.(....).))...((((..((...(((((((((...-)))))))))((((.(((.....)))....))))))..))))))).(((((((.(((...))).))))))). ( -36.60) >sy_sa.0 1670096 118 - 2516575/1360-1480 UUUUAAGCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGAUUGG-AUUGUACGUCCAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGG .......(..((.(....).))...)..........(((((((((...-)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..))).. ( -33.00) >sp_pn.0 1851953 120 + 2038615/1360-1480 UAUGUAGUGAUGGAGGGACGCAGUAGGCUAACUAAAGCAGACGAUUGGAAGAGUCUGUCUAAGCAGUGAGGUGUGAAUUGAGUCAAAUGCUUAAUUCUAUAACAUUGAGCUGUGAUGGGG ..........((((.((((((..(((.....)))..))...(........).)))).)))).(((((..((((((((((((((.....)))))))))....)))))..)))))....... ( -31.20) >sp_ag.0 1905165 120 - 2160267/1360-1480 UAUAUAGUGAUGGAGGGACGCAGUAGGCUAACUAAACCAGACGAUUGGAAGUGUCUGGUCAAACAGUGAGGUGUGAUAUGAGUCAAAUGCUUAUAUCUUUAACAUUGAGCUGUGACGAGG ......((..(((..((((((..(((.....)))..((((....))))..))))))..))).(((((..((((((((((((((.....)))))))))....)))))..))))).)).... ( -31.80) >consensus UUUUAAGCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGAUUGG_AUUGUACGUCUAAGCAGUGAGGUUGAGUAUUAGGCAAAUCCGGAACUC_UUAAGAUUGAGCUGUGAUGGGG .....(((..((.(....).))....))).......(((((((((....)))))))))(((.(((((..(((((((((((.((.....))))))))).....))))..)))))..))).. (-25.12 = -22.20 + -2.92) # Strand winner: reverse (1.00)

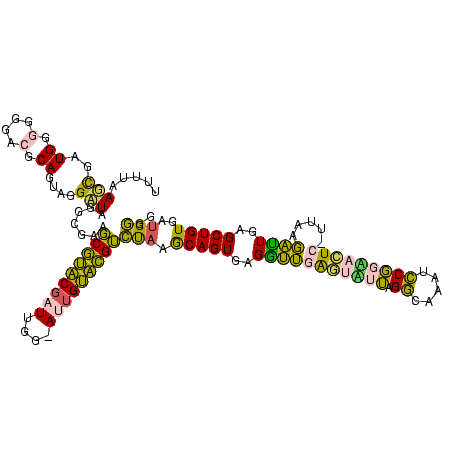
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.92 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -41.62 |
| Energy contribution | -39.62 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.24 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/1480-1600 CCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAGCAUCG .((((.((((((....(((((((...((.....)).)))))))))))))((((((...(((.(.(((....))).).)))...)))).(((((........))))).)).......)))) ( -41.80) >sy_au.0 2275427 120 - 2809422/1480-1600 CCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCG .(((((((((((....(((((((...((.....)).)))))))))))))((((((...(((.(.(((....))).).)))...)))).(((((........))))).))......))))) ( -43.20) >sy_sa.0 1670096 120 - 2516575/1480-1600 CCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAUAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCAUUAUUCG (((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...)))).(((((........))))).............. ( -41.50) >sp_pn.0 1851953 113 + 2038615/1480-1600 CCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUA-------GAG ((((..((((((....(((((((...((.....)).))))))))))))).........(((.(.(((....))).).))).))))...(((((........)))))....-------... ( -39.10) >sp_ag.0 1905165 113 - 2160267/1480-1600 CCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUA-------GAG ((((..((((((....(((((((...((.....)).))))))))))))).........(((.(.(((....))).).))).))))...(((((........)))))....-------... ( -39.10) >consensus CCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAAAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACC_____UCG ......((((((....(((((((...((.....)).)))))))))))))((((((...(((.(.(((....))).).)))...)))).(((((........))))).))........... (-41.62 = -39.62 + -2.00) # Strand winner: reverse (1.00)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -37.92 |
| Consensus MFE | -36.60 |
| Energy contribution | -34.68 |
| Covariance contribution | -1.92 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.719142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/1520-1640 GCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGC (((..((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))................))) ( -38.80) >sy_au.0 2275427 120 - 2809422/1520-1640 GCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGC (((..((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))................))) ( -38.80) >sy_sa.0 1670096 120 - 2516575/1520-1640 GCCUACGCCUAUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGC (((..((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))................))) ( -38.80) >sp_pn.0 1851953 120 + 2038615/1520-1640 GGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGC (((.(.(((....))).).)))...(((...((((((.((((((.(((......))))))))).....))))))...(((.(((((((.......))))))).)))........)))... ( -36.80) >sp_ag.0 1905165 120 - 2160267/1520-1640 GGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGC ((...((((....((..(((....)))..))((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))..............))... ( -36.40) >consensus GCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGC ((....(((....)))....))...((....((((((.((((((.(((......))))))))).....))))))...((..(((((((.......)))))))..)).........))... (-36.60 = -34.68 + -1.92)

| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -19.74 |
| Energy contribution | -17.82 |
| Covariance contribution | -1.92 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/1600-1720 UGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU-AC .(((((((.......)))))))...(((.........)))........(((((((.((....((((((.....)).)))).......)).)))))))....................-.. ( -22.10) >sy_au.0 2275427 119 - 2809422/1600-1720 UGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCA-AA ((((((((.......))).......(((.........)))........(((((((.((....((((((.....)).)))).......)).)))))))...............)))))-.. ( -22.20) >sy_sa.0 1670096 120 - 2516575/1600-1720 UGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUAC .(((((((.......)))))))...(((.........)))........(((((((.((....((((((.....)).)))).......)).)))))))....................... ( -22.10) >sp_pn.0 1851953 118 + 2038615/1600-1720 UGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCACUCCUCACGGUAUACCUUCAUCACACAUAGAACGCUCUCCUACCAUACC--UAUA .(((((((.......)))))))...((.....((((((((((..........))))((....))........)))))).......((.......))..))..............--.... ( -19.70) >sp_ag.0 1905165 120 - 2160267/1600-1720 UGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUGACACUUUU .(((((((.......))))))).(((((((.((((..((........))..)))).))...)))))...(((.((((..(.((((........)))).)......)))).)))....... ( -24.10) >consensus UGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU_AA .(((((((.......)))))))...(((.........)))........(((((((.((....((((((.....)).)))).......)).)))))))....................... (-19.74 = -17.82 + -1.92)

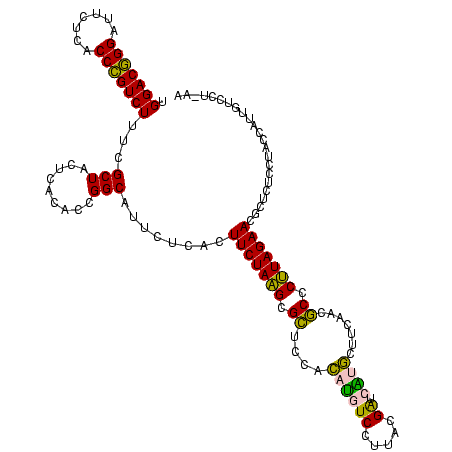
| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -41.28 |
| Energy contribution | -37.72 |
| Covariance contribution | -3.56 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.68 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.41 |
| SVM RNA-class probability | 0.999892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/1600-1720 GU-AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCA ..-...(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))...........((((((.......)))))).. ( -40.90) >sy_au.0 2275427 119 - 2809422/1600-1720 UU-UGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCA ..-...(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))...........((((((.......)))))).. ( -39.90) >sy_sa.0 1670096 120 - 2516575/1600-1720 GUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCA ......(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))...........((((((.......)))))).. ( -40.90) >sp_pn.0 1851953 118 + 2038615/1600-1720 UAUA--GGUAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACAGGUGAGAAUCCUGUCCA (((.--.((((((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....)))).))))..)))......((((((.......)))))).. ( -38.90) >sp_ag.0 1905165 120 - 2160267/1600-1720 AAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGCAAGACAGGUGAGAAUCCUGUCCA ....((((..(((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))).........))))..((((((.......)))))).. ( -41.60) >consensus GU_AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCA ......(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))...........((((((.......)))))).. (-41.28 = -37.72 + -3.56) # Strand winner: reverse (1.00)

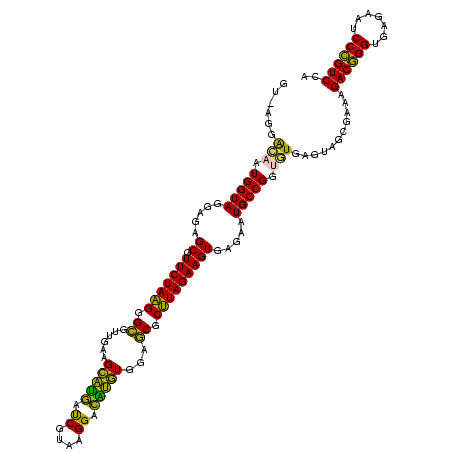
| Location | 2,539,203 – 2,539,321 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -22.25 |
| Consensus MFE | -20.56 |
| Energy contribution | -17.68 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.507408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 118 + 2685015/1640-1760 AUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU-ACGGACAAUCCACAGCUUCGGUAAUAUGUUU-AGCCCCGGUA ........(((((((.((....((((((.....)).)))).......)).)))))))........(((((((((((.-..))))))).....((....)).........-......)))) ( -22.20) >sy_au.0 2275427 118 - 2809422/1640-1760 AUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCA-AAGGACAAUCCACAGCUUCGGUAAUAUGUUU-AGCCCCGGUA ........(((((((.((....((((((.....)).)))).......)).)))))))........(((((((((((.-..))))))).....((....)).........-......)))) ( -22.20) >sy_sa.0 1670096 119 - 2516575/1640-1760 AUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUACGGACAAUCCACAGCUUCGGUAAUAUGUUU-AGCCCCGGUA ........(((((((.((....((((((.....)).)))).......)).)))))))........(((((((((((....))))))).....((....)).........-......)))) ( -23.70) >sp_pn.0 1851953 118 + 2038615/1640-1760 AUUCUCACUUCUAUGCGUUCCAGCACUCCUCACGGUAUACCUUCAUCACACAUAGAACGCUCUCCUACCAUACC--UAUAAAGGUAUCCACAGCUUCGGUAAAUUGUUUUAGCCCCGGUA ..............((((((.............((....)).............)))))).....(((((((((--(....)))))).....((....))................)))) ( -19.37) >sp_ag.0 1905165 120 - 2160267/1640-1760 AUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUGACACUUUUGUGUCAUCCACAGCUUCGGUAAUAUGUUUUAGCCCCGGUA ........((((((((((....)))).......((....))..........))))))........(((((((((((....))))))).....((....))................)))) ( -23.80) >consensus AUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU_AAGGACAAUCCACAGCUUCGGUAAUAUGUUU_AGCCCCGGUA ........(((((((.((....((((((.....)).)))).......)).)))))))........(((((((((((....)))))))..........(((...........)))..)))) (-20.56 = -17.68 + -2.88)

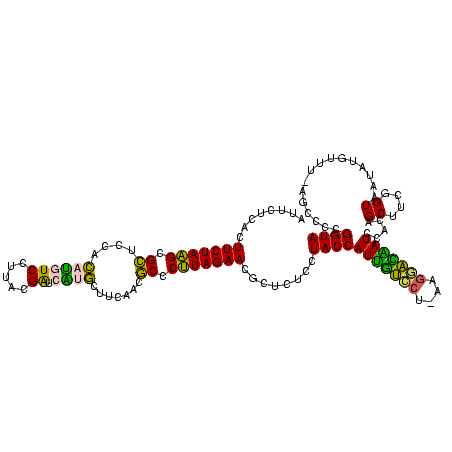
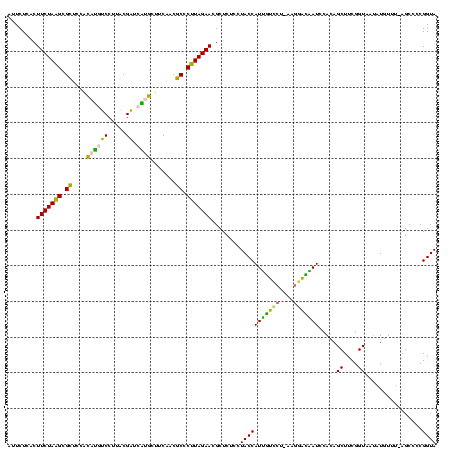
| Location | 2,539,203 – 2,539,321 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.44 |
| Mean single sequence MFE | -37.99 |
| Consensus MFE | -40.86 |
| Energy contribution | -36.22 |
| Covariance contribution | -4.64 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.89 |
| Structure conservation index | 1.08 |
| SVM decision value | 3.43 |
| SVM RNA-class probability | 0.999200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 118 + 2685015/1640-1760 UACCGGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCGU-AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAU ..(((.((((-..............)))).)))(((((((..-.))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))...... ( -36.34) >sy_au.0 2275427 118 - 2809422/1640-1760 UACCGGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCUU-UGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAU ..(((.((((-..............)))).)))(((((((..-.))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))...... ( -35.34) >sy_sa.0 1670096 119 - 2516575/1640-1760 UACCGGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCGUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAU ..(((.((((-..............)))).)))(((((((....))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))...... ( -40.64) >sp_pn.0 1851953 118 + 2038615/1640-1760 UACCGGGGCUAAAACAAUUUACCGAAGCUGUGGAUACCUUUAUA--GGUAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAU ..(((.((((...............)))).)))((((((....)--))))).........((.((((((((((......(((((.((....)).)))))...))))))))))))...... ( -36.66) >sp_ag.0 1905165 120 - 2160267/1640-1760 UACCGGGGCUAAAACAUAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAU ..(((.((((...............)))).)))(((((((....))))))).........((.((((((((((......(((((.((....)).)))))...))))))))))))...... ( -40.96) >consensus UACCGGGGCU_AAACAUAUUACCGAAGCUGUGGAUUGUCCGU_AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAU ..(((.((((...............)))).)))(((((((....))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))...... (-40.86 = -36.22 + -4.64) # Strand winner: reverse (1.00)



| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -29.16 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/1760-1880 CAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGA-UGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCUUUUCCACUUAACAUAU .......((((...((((...(((((((.((((......))..........((.-(((((.......))))).)))))))))))....)))).))))....................... ( -31.40) >sy_au.0 2275427 119 - 2809422/1760-1880 CAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGA-UGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCUUUUCCACUUAACAUAU .......((((...((((...(((((((.((((......))..........((.-(((((.......))))).)))))))))))....)))).))))....................... ( -31.40) >sy_sa.0 1670096 119 - 2516575/1760-1880 CAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGA-UGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCUUUUCCACUUAACAUAU .......((((...((((...(((((((.((((......))..........((.-(((((.......))))).)))))))))))....)))).))))....................... ( -31.40) >sp_pn.0 1851953 120 + 2038615/1760-1880 CAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAUU .......((....(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..)))).....))................. ( -25.80) >sp_ag.0 1905165 120 - 2160267/1760-1880 CAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAUAU .......((....(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..)))).....))................. ( -25.80) >consensus CAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGA_UGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCUUUUCCACUUAACAUAU .......((((...(((((..(((((((.((((......))..............(((((.......)))))...)))))))))..)).))).))))....................... (-24.02 = -24.34 + 0.32)

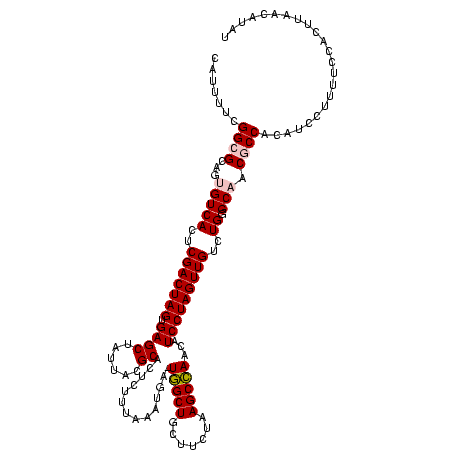
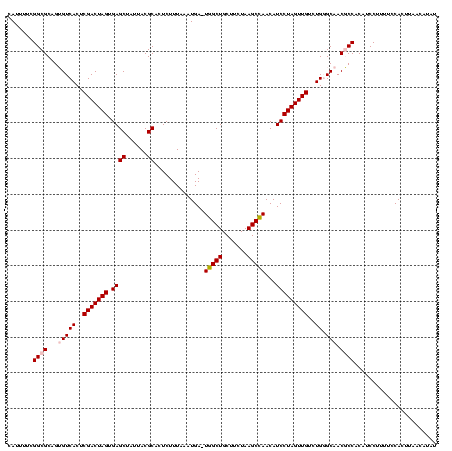
| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.82 |
| Mean single sequence MFE | -33.14 |
| Consensus MFE | -28.80 |
| Energy contribution | -28.08 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/1760-1880 AUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAACUAGGAUGUUGGCUUAGAAGCAGCCA-UCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUG .................((.((..(.((..(...(((.....(((((.(((((.......)))))-.)))))....((((.((....)).)))).)))..)..)).)..))))....... ( -30.90) >sy_au.0 2275427 119 - 2809422/1760-1880 AUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAACUAGGAUGUUGGCUUAGAAGCAGCCA-UCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUG .................((.((..(.((..(...(((.....(((((.(((((.......)))))-.)))))....((((.((....)).)))).)))..)..)).)..))))....... ( -30.90) >sy_sa.0 1670096 119 - 2516575/1760-1880 AUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAACUAGGAUGUUGGCUUAGAAGCAGCCA-UCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUG .................((.((..(.((..(...(((.....(((((.(((((.......)))))-.)))))....((((.((....)).)))).)))..)..)).)..))))....... ( -30.90) >sp_pn.0 1851953 120 + 2038615/1760-1880 AAUUGUUAAGUGGAAAAGGAUGUGGGGUUGCACAGACAACUAGGAUGUUAGCUUAGAAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUG .................((.((..((((..(...(((.....(((((.(((((.......)))))..)))))....((((.((....)).)))).)))..)..))))..))))....... ( -36.50) >sp_ag.0 1905165 120 - 2160267/1760-1880 AUAUGUUAAGUGGAAAAGGAUGUGGGGUUGCACAGACAACUAGGAUGUUAGCUUAGAAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUG .................((.((..((((..(...(((.....(((((.(((((.......)))))..)))))....((((.((....)).)))).)))..)..))))..))))....... ( -36.50) >consensus AUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAACUAGGAUGUUGGCUUAGAAGCAGCCA_UCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUG .................((.((..(.((..(...(((.....(((((..((((.......))))...)))))....((((.((....)).)))).)))..)..)).)..))))....... (-28.80 = -28.08 + -0.72) # Strand winner: reverse (1.00)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.58 |
| Mean single sequence MFE | -33.64 |
| Consensus MFE | -30.80 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/1840-1960 AACUUGGGAGUCAGAACACGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAAC .........(((.(((((((((........)))))))))....(((.(((.(((((..(((((......))).(((.(((....)))...)))....)).)).)))))).))).)))... ( -36.20) >sy_au.0 2275427 120 - 2809422/1840-1960 AACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAAC .........(((.(((((((((........)))))))))....(((.(((.(((((..(((((......))).(((.(((....)))...)))....)).)).)))))).))).)))... ( -34.40) >sy_sa.0 1670096 120 - 2516575/1840-1960 AACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAAC .........(((.(((((((((........)))))))))....(((.(((.(((((..(((((......))).(((.(((....)))...)))....)).)).)))))).))).)))... ( -35.80) >sp_pn.0 1851953 120 + 2038615/1840-1960 UGGUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAAUUGUUAAGUGGAAAAGGAUGUGGGGUUGCACAGACAAC ..(((.(((..(.(((((((..(......)..)))))))....(((......)))...((((...((....((((..(((....)))..).)))...))..)))))..)))...)))... ( -32.60) >sp_ag.0 1905165 120 - 2160267/1840-1960 UAAUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUGUGGGGUUGCACAGACAAC .........(((.(((((((..(......)..)))))))..........((((((.((((.........))))........((((((...(....)..))))))))))))....)))... ( -29.20) >consensus AACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUGUGGCGUUGCCCAGACAAC .........(((.((((((((((......))))))))))....(((...(((((((.((((((......))).(((.(((....)))...)))....)).).))).))))))).)))... (-30.80 = -30.00 + -0.80) # Strand winner: reverse (0.96)

| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -26.32 |
| Energy contribution | -25.92 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/1880-2000 GGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACACGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAU ((..((((.((((.((.....)).))))..)............((((..(((.(((((((((........)))))))))....(((......))).)))..))))....))).))..... ( -33.20) >sy_au.0 2275427 120 - 2809422/1880-2000 GGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAU ((..((((.((((.((.....)).))))..)............((((..(((.(((((((((........)))))))))....(((......))).)))..))))....))).))..... ( -31.40) >sy_sa.0 1670096 120 - 2516575/1880-2000 GGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUCAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAU ((..((((.((((.((.....)).))))..)............((((..(((.(((((((((........)))))))))....(((......))).)))..))))....))).))..... ( -32.80) >sp_pn.0 1851953 120 + 2038615/1880-2000 GAUUACCAAUCUCAGAUAAACUCCGAAUGCCAAUGAAUUAUGGUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAU ((((((..((((.((....((((((...((((........))))))((((.....)))))))).)).))))..))))))....(((......))).((((.........))))....... ( -32.10) >sp_ag.0 1905165 120 - 2160267/1880-2000 GAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAGAUAUAAUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAU ((((((..((((.((....((((..........(((.......)))((((.....)))))))).)).))))..))))))....(((......))).((((.........))))....... ( -31.00) >consensus GGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUGAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAU ....(((..((((.((.....)).)))).................((..(((.((((((((((......))))))))))....(((......))).)))..))......)))........ (-26.32 = -25.92 + -0.40) # Strand winner: forward (0.99)

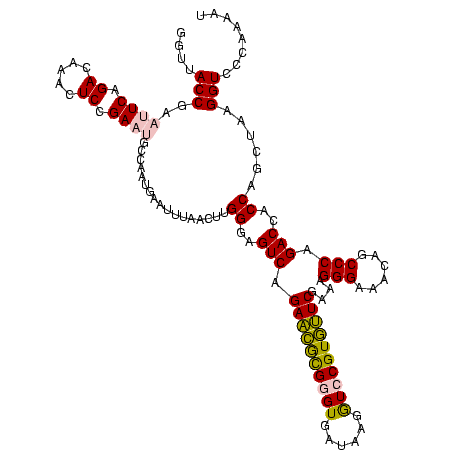
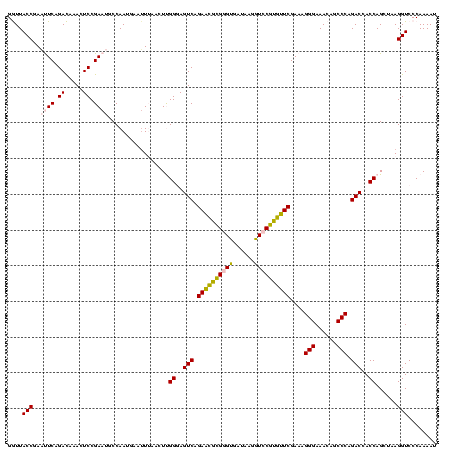
| Location | 2,539,203 – 2,539,321 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -35.06 |
| Consensus MFE | -31.64 |
| Energy contribution | -29.88 |
| Covariance contribution | -1.76 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.540805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 118 + 2685015/2040-2160 AACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAA--GUGAUGAUU ..(((.(((((........))))))))....(((..((((..((((((...(((((......)))))((........))))))))...(((((....)))))))))..)--))....... ( -33.90) >sy_au.0 2275427 118 - 2809422/2040-2160 AACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAA--GUGAUGAUU ..(((.(((((........))))))))....(((((((.((((((((...)))).......(((((((.((..(((......)))...)).))))))))))).)))).)--))....... ( -38.50) >sy_sa.0 1670096 118 - 2516575/2040-2160 AACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAA--GUGAUGAUU ..(((.(((((........))))))))((.(.(((.((.((((((((...)))).......(((((((.((..(((......)))...)).))))))))))).)).)))--.).)).... ( -36.70) >sp_pn.0 1851953 120 + 2038615/2040-2160 AACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCGUCGACAUUAGAGAUU ..(((((.(((........))))))))....((.(((....(((((((...(((((......)))))((........)))))))....(((((....))))))).....))).))..... ( -31.80) >sp_ag.0 1905165 119 - 2160267/2040-2160 AACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCGUCGAUGUUA-AGUCU ..(((((.(((........))))))))..((((((.(((.(.(((.....((((((......))))))....))).).)))..(((..(((((....)))))...)))....))-)))). ( -34.40) >consensus AACCGACUUACGUUGAAAAGUGAGCGGAUGAAUUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAA__GUGAUGAUU ..(((.(((((........))))))))......((.((.((((((((...)))).......(((((((.((..(((......)))...)).))))))))))).)).))............ (-31.64 = -29.88 + -1.76) # Strand winner: reverse (1.00)

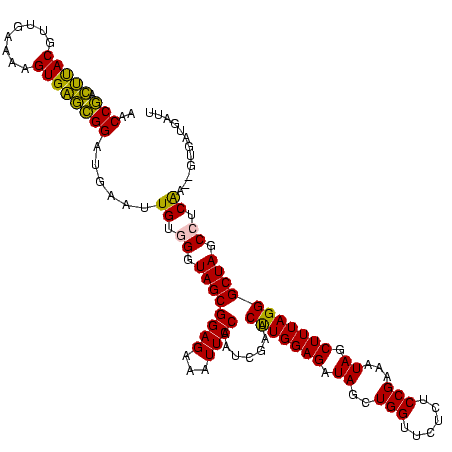
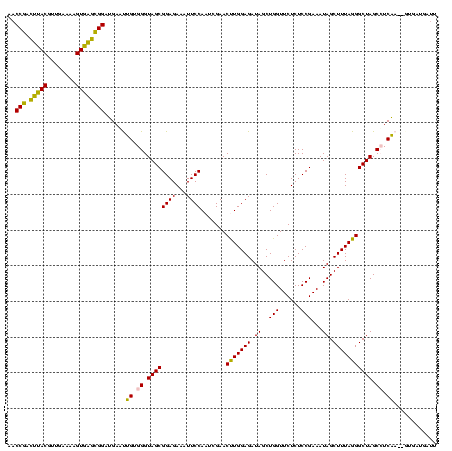
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -24.84 |
| Consensus MFE | -19.60 |
| Energy contribution | -17.36 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2080-2200 AGAACCAGCUAUCUCCAAGUUCGAUUGGAAUUUCUCCGCUACCCACAGUUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGACCA .((((((((....(((((......)))))........(((......)))......))).........((....)).)))))((((.....(((((......))))).........)))). ( -22.74) >sy_au.0 2275427 120 - 2809422/2080-2200 AGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCUCAGUUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGACCA .((((((((....(((((......)))))........(((......)))......))).........((....)).)))))((((.....(((((......))))).........)))). ( -22.24) >sy_sa.0 1670096 120 - 2516575/2080-2200 AGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACAACUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGGUCA ..............(((((((.((.((((......(((..(((.((...........................)).))).)))...))))(((((......)))))..)))))))))... ( -22.73) >sp_pn.0 1851953 120 + 2038615/2080-2200 AGAACCAGCUAUCUCCAAGUUCGUUUGGAAUUUCUCCGCUACCCACAAGUCAUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGCGUCUUACCGCACCUUCAACCUGCUCA .(((((((.....((((((....))))))............................((((........)))).)))))))(((......(((((......))))).....)))...... ( -28.90) >sp_ag.0 1905165 120 - 2160267/2080-2200 AGAACCAGCUAUCUCCAAGUUCGUUUGGAAUUUCUCCGCUACCCACAAGUCAUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGAGUUUUACCUCACCUUCAACCUGCUCA .(((((((.....((((((....))))))............................((((........)))).)))))))(((......(((((......))))).....)))...... ( -27.60) >consensus AGAACCAGCUAUCUCCAAGUUCGAUUGGAAUUUCUCCGCUACCCACAAGUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGAUCA .(((((((((.......)))).((.((((.......................)))).)).................)))))(((......(((((......))))).....)))...... (-19.60 = -17.36 + -2.24)


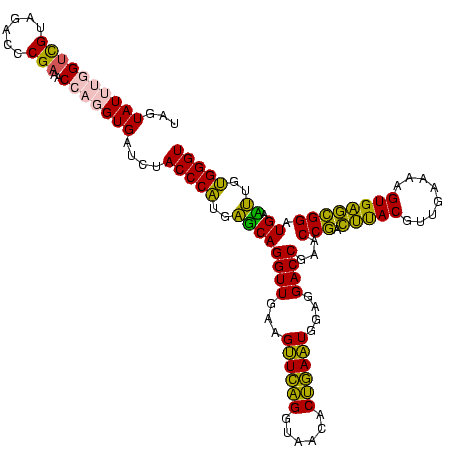
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.08 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -34.94 |
| Energy contribution | -30.62 |
| Covariance contribution | -4.32 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.24 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2080-2200 UGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCU .((((........((((((......))))))....))))((((((.(((((........)))))).......((((.....)))).....((((((......))))))......))))). ( -31.70) >sy_au.0 2275427 120 - 2809422/2080-2200 UGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCU .(((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)))...(((.(.(((....)))).)))(((((.((......))))))). ( -32.30) >sy_sa.0 1670096 120 - 2516575/2080-2200 UGACCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCU ...(((((((((.((((((......))))))((((..((...(((.(((((........))))))))....((((....)))))).....))))..)).))))))).............. ( -34.90) >sp_pn.0 1851953 120 + 2038615/2080-2200 UGAGCAGGUUGAAGGUGCGGUAAGACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCU ......((((...((((((......))))))....))))((((((((((((........)))))..........................((((((......))))))....))))))). ( -37.70) >sp_ag.0 1905165 120 - 2160267/2080-2200 UGAGCAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCU ......((((...((((((......))))))....))))((((((((((((........)))))..........................((((((......))))))....))))))). ( -36.40) >consensus UGAGCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAAUUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCU .(((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))).........((((...))))....(((((...........))))). (-34.94 = -30.62 + -4.32) # Strand winner: reverse (1.00)

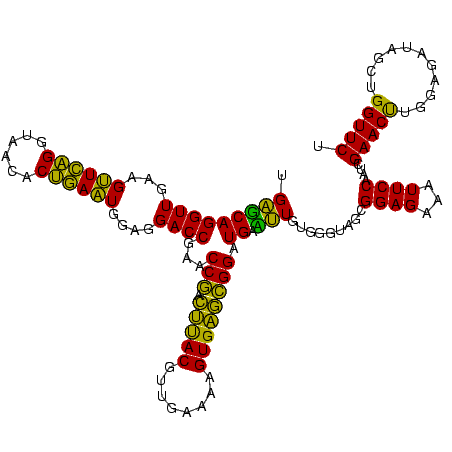
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.08 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -41.82 |
| Energy contribution | -38.66 |
| Covariance contribution | -3.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.33 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2120-2240 GAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGU ((.((((((((((......)))..))))))).)).(((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))))))))) ( -42.30) >sy_au.0 2275427 120 - 2809422/2120-2240 UAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCUUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGU ...((((((((((......)))..)))))))....(((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))) ( -41.20) >sy_sa.0 1670096 120 - 2516575/2120-2240 GAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGACCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGU ((.((((((((((......)))..))))))).)).(((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))) ( -42.50) >sp_pn.0 1851953 120 + 2038615/2120-2240 UAGUAUCAUGACGUAGACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAGACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGU ..((..((((.((......))....))))..))..(((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))) ( -39.30) >sp_ag.0 1905165 120 - 2160267/2120-2240 UAGUAUCAUGUUGUAGACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGU .....((((((.((.........)).))))))...(((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))) ( -39.20) >consensus UAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGAGCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAAUUGUGGGU ...((((((((((......)))..)))))))....(((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))) (-41.82 = -38.66 + -3.16) # Strand winner: reverse (1.00)


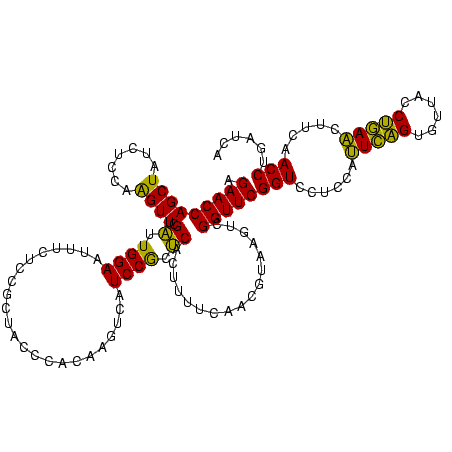
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -39.23 |
| Consensus MFE | -37.82 |
| Energy contribution | -37.02 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2320-2440 ACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAGAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAU .......((....))(((((((...(((((((((....)))((((........))))........)))))).......((((..(((((....)))))..))))..)))))))....... ( -40.00) >sy_au.0 2275427 120 - 2809422/2320-2440 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAU .......((....))(((((((...(((((((((....)))).((........))...........))))).......((((..(((((....)))))..))))..)))))))....... ( -39.50) >sy_sa.0 1670096 120 - 2516575/2320-2440 ACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAUAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAU .......((....))(((((((...(((((((((....)))((((........))))........)))))).......((((..(((((....)))))..))))..)))))))....... ( -40.00) >sp_pn.0 1851953 120 + 2038615/2320-2440 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAU ....(((((....)((..((((.....((.((((....)))).))........))))....))..))))((((((((.((((..(((((....)))))..)))).))....))))))... ( -38.32) >sp_ag.0 1905165 120 - 2160267/2320-2440 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAU ....(((((....)((..((((.....((.((((....)))).))........))))....))..))))((((((((.((((..(((((....)))))..)))).))....))))))... ( -38.32) >consensus ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAU .......((....))(((((((...(((((((((....)))).((........))...........))))).......((((..(((((....)))))..))))..)))))))....... (-37.82 = -37.02 + -0.80) # Strand winner: reverse (0.99)

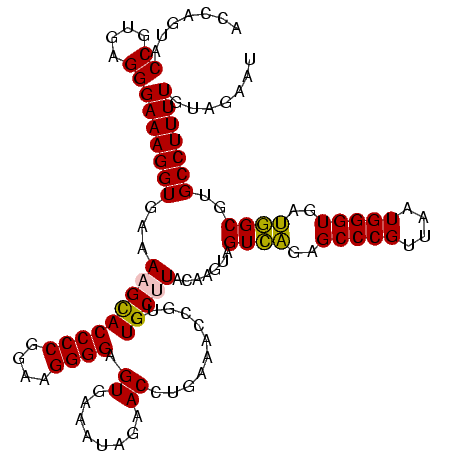
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -34.88 |
| Energy contribution | -33.12 |
| Covariance contribution | -1.76 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2400-2520 ACGACGGAGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGG .((((((((.........))))))))...((((((((((.....))))...(((...((((.((........)).)))).(((....((....))....)))....)))..))))))... ( -35.30) >sy_au.0 2275427 120 - 2809422/2400-2520 ACGACGGAGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGG .((((((((.........))))))))...((((((((((.....))))...(((...((((.((........)).)))).(((....((....))....)))....)))..))))))... ( -37.30) >sy_sa.0 1670096 120 - 2516575/2400-2520 ACGACGGAACACGAGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCCAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGG .((((((((.........)))))))).....((((((......))))))..(((...((((.((........)).)))).(((....((....))....)))....)))...((....)) ( -35.30) >sp_pn.0 1851953 120 + 2038615/2400-2520 ACGGCGGGACACGUGAAAUCCCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCCCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGG .((((((((.........)))))))).....((((((......))))))..........(((((.......................((....))....((((.....))))..))))). ( -41.50) >sp_ag.0 1905165 120 - 2160267/2400-2520 ACGGCGAGACACGCGAAAUCUCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGG .((((((((.........)))))))).....((((((......)))))).(((....((((.((........)).))))........((....))....((((.....))))..)))... ( -36.70) >consensus ACGACGGAACACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCCAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGG .((((((((.........)))))))).....((((((......))))))........((((....)))).(((...(((.(((....((....))....)))......)))..))).... (-34.88 = -33.12 + -1.76) # Strand winner: forward (0.56)

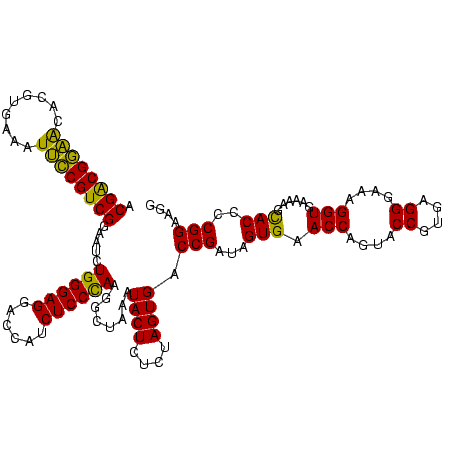

| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -29.04 |
| Energy contribution | -25.80 |
| Covariance contribution | -3.24 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/2480-2600 GAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGG-AUCCUGAGUACGACGGAGCACGUGAAAUUCCGUCGGAAUCUGGGAGGAC ...(((.((....)).))).((((...(((.......)))....)))).....(((((......))))).-.((((((...((((((((.........))))))))...)).)))).... ( -28.50) >sy_au.0 2275427 119 - 2809422/2480-2600 GAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUCUCUCUUGAGUGG-AUCCUGAGUACGACGGAGCACGUGAAAUUCCGUCGGAAUCUGGGAGGAC ((.((((((....)))))).)).....(((.......)))...(((...........(((....)))...-.((((((...((((((((.........))))))))...)).)))).))) ( -32.10) >sy_sa.0 1670096 119 - 2516575/2480-2600 GAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCACUCUUGAGUGG-AUCCUGAGUACGACGGAACACGAGAAAUUCCGUCGGAAUCUGGGAGGAC ((.((((((....)))))).)).....(((.......)))...(((.........((((((....)))))-)((((((...((((((((.........))))))))...)).)))).))) ( -35.90) >sp_pn.0 1851953 119 + 2038615/2480-2600 GAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGU-CUUUGUACUUAGCAGUAUCCUGAGUACGGCGGGACACGUGAAAUCCCGUCGGAAUCUGGGAGGAC (..((((((....))))))..)...(((.((....)))))............((-((((((((((((.......))))))))(((((((.........))))))).........)))))) ( -32.30) >sp_ag.0 1905165 120 - 2160267/2480-2600 GAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGUACGGCGAGACACGCGAAAUCUCGUCGGAAUCUGGGAGGAC .......(((..(((((........(((.((....))))).((((......)))).....)))))..)))..((((((...((((((((.........))))))))...)).)))).... ( -29.00) >consensus GAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAGUCUCUCUUGAGUGG_AUCCUGAGUACGACGGAACACGUGAAAUUCCGUCGGAAUCUGGGAGGAC ...((((((....)))))).((.....(((.......)))................................((((((...((((((((.........))))))))...)).)))))).. (-29.04 = -25.80 + -3.24) # Strand winner: reverse (1.00)

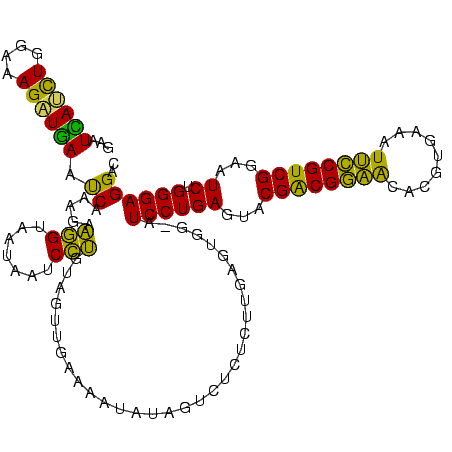
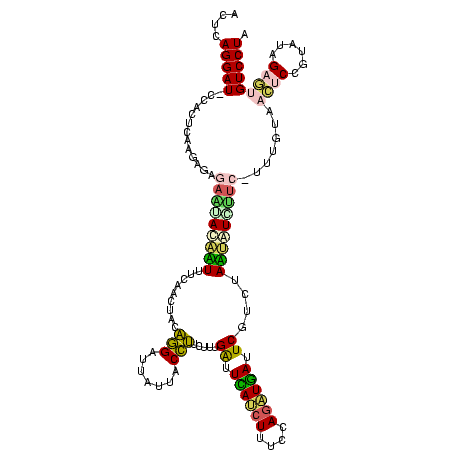
| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -24.38 |
| Consensus MFE | -15.16 |
| Energy contribution | -14.60 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/2520-2640 ACUCAGGAU-CCACUCAAGAGAGAAUAUAUUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUA ....((((.-.(((((.((((.(((((((((..........((((((..........))))))........(((((...))))))))))))))))))(((......))).))))))))). ( -23.20) >sy_au.0 2275427 119 - 2809422/2520-2640 ACUCAGGAU-CCACUCAAGAGAGACAACAUUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUA ....((((.-.(((((.((((.(((.(((((...(((...(((.......))).....((.((((((....)))))).))))).))))).)))))))(((......))).))))))))). ( -29.70) >sy_sa.0 1670096 119 - 2516575/2520-2640 ACUCAGGAU-CCACUCAAGAGUGAACAAACUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUA ....((((.-.(((((......(((((.(((...(((...(((.......))).....((.((((((....)))))).))))).))).)))))...((((......))))))))))))). ( -28.40) >sp_pn.0 1851953 112 + 2038615/2520-2640 ACUCAGGAUACUGCUAAGUACAAAG-ACUAUUUUAAAUACGAGGCUAUUACUCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUAUAAUCUU------UGAGUCCACAUUGCA-GUCCUA ....((((..((((......(((((-(.(((.........(((.......)))..(((((.........)))))..........))).))))------)).(....)...)))-))))). ( -17.50) >sp_ag.0 1905165 113 - 2160267/2520-2640 ACUCAGGAUACUGCUAAGGUUAAUCUAUCAUUUUAAAUACGAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUA ....((((..((((..(((((((((...............(((.......))).....(..((((((....))))))..).........)))------))).))).....)))-))))). ( -23.10) >consensus ACUCAGGAU_CCACUCAAGAGAGAAUACAAUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUAUCUUC_UUUGUAACUCCGUAUAGAGUGUCCUA ....(((((.............(((((((((.........(((.......))).....((.((((((....)))))).))....))))))))).......((((......))))))))). (-15.16 = -14.60 + -0.56)

| Location | 2,539,203 – 2,539,322 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Mean single sequence MFE | -33.09 |
| Consensus MFE | -23.10 |
| Energy contribution | -20.66 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.62 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.04 |
| SVM RNA-class probability | 0.999772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 119 + 2685015/2520-2640 UAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGG-AUCCUGAGU ((((((((((.....((((.......((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...)))))))))))))).))))).-.)))))... ( -29.21) >sy_au.0 2275427 119 - 2809422/2520-2640 UAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUCUCUCUUGAGUGG-AUCCUGAGU ((((((((((.....((((.......((((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))))))).))))).-.)))))... ( -39.01) >sy_sa.0 1670096 119 - 2516575/2520-2640 UAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCACUCUUGAGUGG-AUCCUGAGU ((((((((((.....(((((.......(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))))))).))))).-.)))))... ( -39.31) >sp_pn.0 1851953 112 + 2038615/2520-2640 UAGGAC-UGCAAUGUGGACUCA------AAGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGU-CUUUGUACUUAGCAGUAUCCUGAGU ((((((-(((...(((....((------((((((((....(..((((((....))))))..)...(((.((....))))).........)))))-))))))))...))))..)))))... ( -27.80) >sp_ag.0 1905165 113 - 2160267/2520-2640 UAGGAC-UGCGAUAUAGGAUUA------AUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGU ((((((-(((.....(((.(((------((((((((....(..((((((....))))))..)...(((.((....))))).........))))).)))))))))..))))..)))))... ( -30.10) >consensus UAGGACACUCUAUACGGAGUUACAAA_GAACAUAAUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAGUCUCUCUUGAGUGG_AUCCUGAGU (((((.((((......)))).......(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))..............)))))... (-23.10 = -20.66 + -2.44) # Strand winner: reverse (1.00)

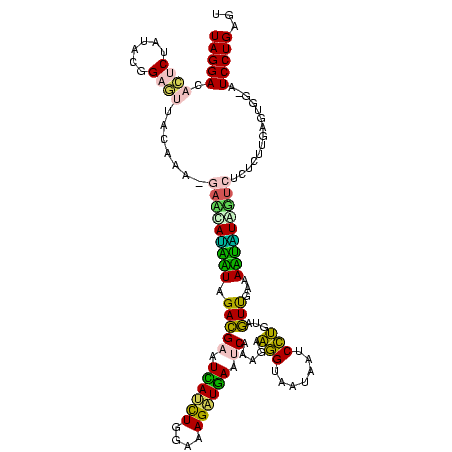
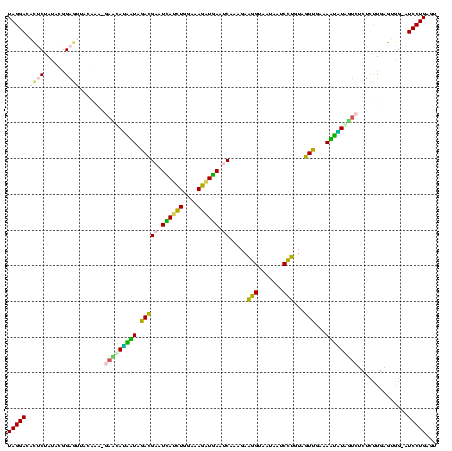
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.10 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -21.32 |
| Energy contribution | -19.48 |
| Covariance contribution | -1.84 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2560-2680 AGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCG .(((..............((.(((.((....)).))).))((((((.........(((((((((......))))....)))))..((((((((....)))))))).))))))...))).. ( -24.30) >sy_au.0 2275427 120 - 2809422/2560-2680 AGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCA .(((..............((.((((((....)))))).))(((((((((.(.((((((((......))))))).).)..)))...((((((((....)))))))).))))))...))).. ( -31.10) >sy_sa.0 1670096 120 - 2516575/2560-2680 AGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUUCCCG .(((..............((.((((((....)))))).))((((((.........(((((((((......))))....)))))..(((((.((....)).))))).))))))...))).. ( -26.40) >sp_pn.0 1851953 113 + 2038615/2560-2680 GAGGCUAUUACUCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUAUAAUCUU------UGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCG ..(((.............(((((...........(((...............------)))..........))-)))........((((((((....))))))))...........))). ( -23.36) >sp_ag.0 1905165 113 - 2160267/2560-2680 GAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCG ((((..((((((......((....))......)))))).)))).........------............(((-(.....(((..((((((((....)))))))).))).....)))).. ( -26.10) >consensus AGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUAUCUUC_UUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUCGUUGGUUUGGGCUCUUCCCG (((((.............((.((((((....)))))).))....................((((......)))))))))......((((((((....))))))))...(((.....))). (-21.32 = -19.48 + -1.84)

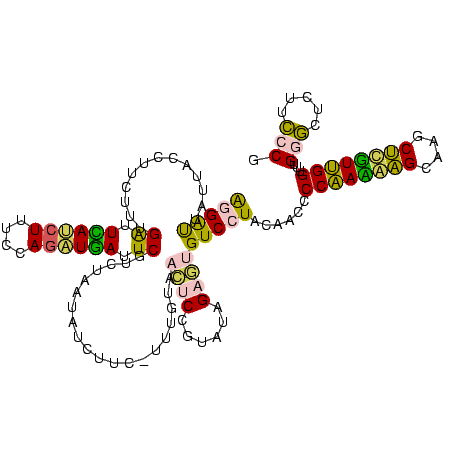
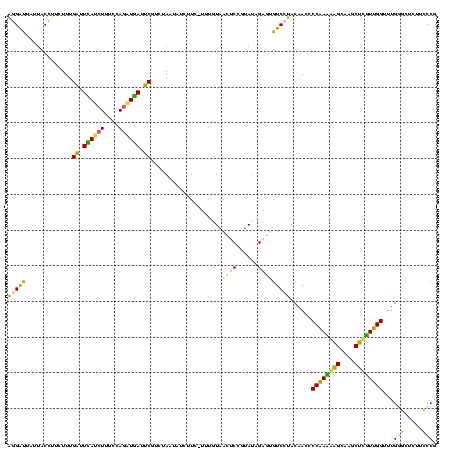
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.10 |
| Mean single sequence MFE | -32.86 |
| Consensus MFE | -28.06 |
| Energy contribution | -25.82 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2560-2680 CGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCU .(((.....)))...((((((((....))))))))..(((((....(((((....))))))))))((((.(((.((....((.(((.((....)).))).))....))....))).)))) ( -27.80) >sy_au.0 2275427 120 - 2809422/2560-2680 UGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCU ((((.....))))..((((((((....)))))))).(((((.....(((((....)))))........))))).......((.((((((....)))))).)).....(((.......))) ( -35.52) >sy_sa.0 1670096 120 - 2516575/2560-2680 CGGGAAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCU .(((.....)))...((((((((....))))))))...((((((....)))))).((((((((...((......))....((.((((((....)))))).)).......)))))..))). ( -33.30) >sp_pn.0 1851953 113 + 2038615/2560-2680 CGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCA------AAGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUC .(((..(((..((..((((((((....)))))))).((((((....-)))))).))..))).------...............((((((....)))))))))...(((.((....))))) ( -34.40) >sp_ag.0 1905165 113 - 2160267/2560-2680 CGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUA------AUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUC ..((((....((((.((((((((....))))))))..))))....)-)))..(((((((...------.)).)))))...(..((((((....))))))..)...(((.((....))))) ( -33.30) >consensus CGGGAAGAGCCCAAACCAAAGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAA_GAACAUAAUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCU .(((.....)))...((((((((....))))))))((((((.....((((......))))....................((.((((((....)))))).))..........)))))).. (-28.06 = -25.82 + -2.24) # Strand winner: reverse (1.00)

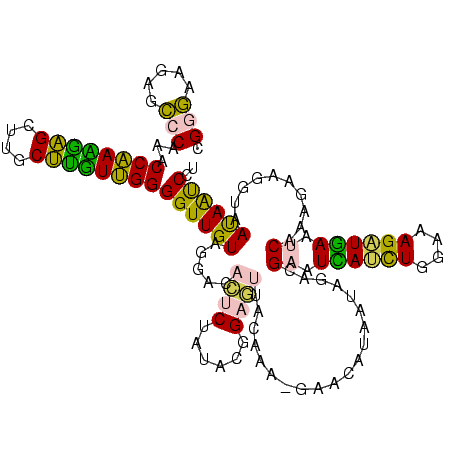
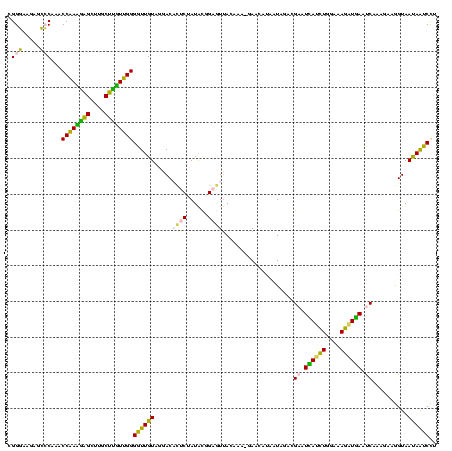
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -27.00 |
| Consensus MFE | -21.38 |
| Energy contribution | -18.62 |
| Covariance contribution | -2.76 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829732 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2600-2720 GUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUC ((((((.........(((((((((......))))....)))))..((((((((....)))))))).))))))..(((((.....((.....))......)))))................ ( -26.00) >sy_au.0 2275427 120 - 2809422/2600-2720 GUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCAUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUUCUUUCUC ........((((((((((((....((....(((((..........((((((((....))))))))..((((.....))))...)))))..)).))).))))))..)))............ ( -29.30) >sy_sa.0 1670096 120 - 2516575/2600-2720 GUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUC (...((((.(((((((.(((....((....(((((....((....(((((.((....)).)))))...(((.....)))))..)))))..)).)))..))))))).))))...)...... ( -24.70) >sp_pn.0 1851953 113 + 2038615/2600-2720 UUCUAUAAUCUU------UGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUC ............------.((((....((.(((-(.....(((..((((((((....)))))))).))).....)))).))...))))(((........)))....((....))...... ( -28.50) >sp_ag.0 1905165 113 - 2160267/2600-2720 UUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUC .........(((------(...........(((-(.....(((..((((((((....)))))))).))).....))))..........(((........)))))))((....))...... ( -26.50) >consensus GUCUAAUAUCUUC_UUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAUUUUUCUUUCUC ..............................(((.......(((..((((((((....)))))))).)))......((((.....((.....))......))))........)))...... (-21.38 = -18.62 + -2.76)

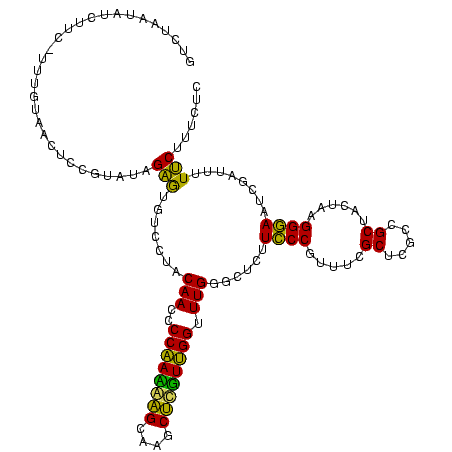
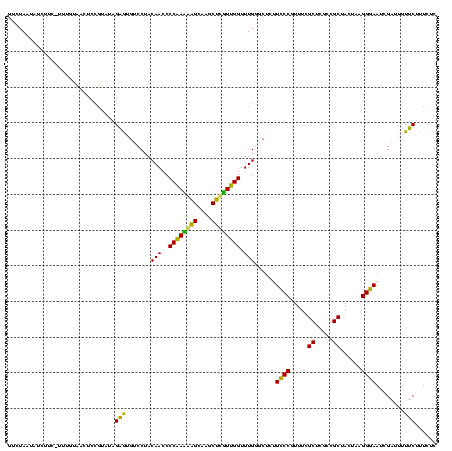
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -25.96 |
| Energy contribution | -22.84 |
| Covariance contribution | -3.12 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2600-2720 GAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGAC ..............((((((..((((..(.((((.(.....(((.....)))...((((((((....))))))))).))))..)..)))).....))))))................... ( -28.80) >sy_au.0 2275427 120 - 2809422/2600-2720 GAGAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAAUGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGAC ............(((....((((.(((((..(((((....((((.....))))..((((((((....)))))))).)))(((((....))))).))..))))).)))).)))........ ( -32.50) >sy_sa.0 1670096 120 - 2516575/2600-2720 GAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGAC ..............((((((..((((..(.((((.(.....(((.....)))...((((((((....))))))))).))))..)..)))).....))))))................... ( -30.40) >sp_pn.0 1851953 113 + 2038615/2600-2720 GAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCA------AAGAUUAUAGAA (((...(((....((.(((((........))))).)).....((((....((((.((((((((....))))))))..))))....)-)))..)))...))).------............ ( -37.70) >sp_ag.0 1905165 113 - 2160267/2600-2720 GAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUA------AUCAUUAUAGAA ..((.........((.(((((........))))).)).....((((....((((.((((((((....))))))))..))))....)-)))............------.))......... ( -32.60) >consensus GAGAAAGAAAAAUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAAGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAA_GAACAUAAUAGAC ......((.....(..(((((......((.....)).....)))))..)..(((.((((((((....))))))))..)))........)).............................. (-25.96 = -22.84 + -3.12) # Strand winner: reverse (1.00)

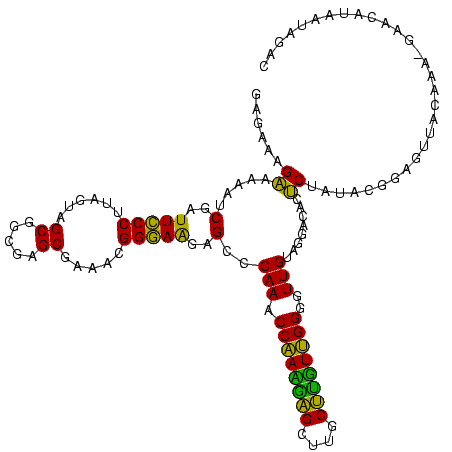
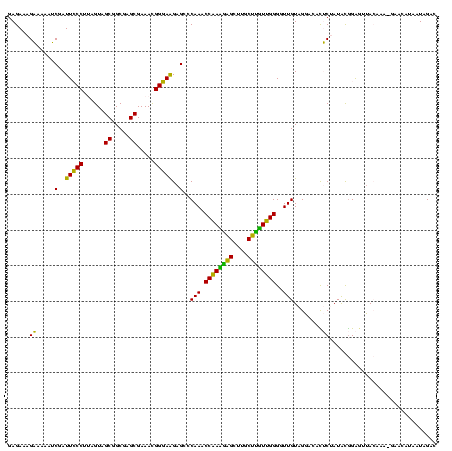
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.00 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -32.72 |
| Energy contribution | -29.52 |
| Covariance contribution | -3.20 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2640-2760 GCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUG .(((..((((.(.(((((......))))).))))).((((..((........))..)))))))....((.....))..((((((.....)))...((((((((....)))))))).))). ( -34.90) >sy_au.0 2275427 120 - 2809422/2640-2760 GCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAAUGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUG ((((..((((.(.(((((......))))).))))).((((..(((......)))..))))....)).))..(((.(....((((.....))))..((((((((....))))))))).))) ( -37.60) >sy_sa.0 1670096 120 - 2516575/2640-2760 GCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUG .(((..((((.(.(((((......))))).))))).((((..((........))..)))))))....((.....))..((((((.....)))...((((((((....)))))))).))). ( -37.20) >sp_pn.0 1851953 120 + 2038615/2640-2760 GAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUG ......(((((..(((..........))).)))))...........((.....((.(((((........))))).)).....)).....(((...((((((((....)))))))).))). ( -35.30) >sp_ag.0 1905165 120 - 2160267/2640-2760 GAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUG ......(((((..(((..........))).)))))...........((.....((.(((((........))))).)).....)).....(((...((((((((....)))))))).))). ( -35.10) >consensus GCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAAGAGCUUGCUUGUUGGGGUUG ......((((...(((((......)))))..))))..................(..(((((......((.....)).....)))))..)..(((.((((((((....))))))))..))) (-32.72 = -29.52 + -3.20) # Strand winner: reverse (1.00)

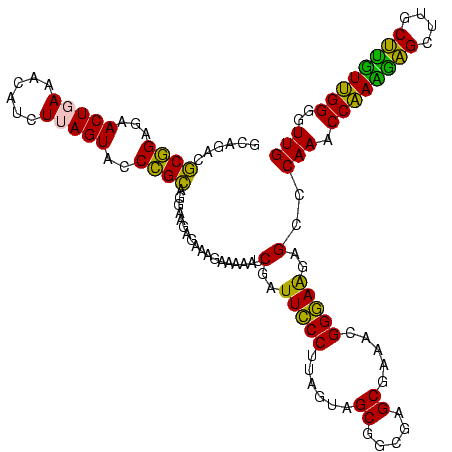
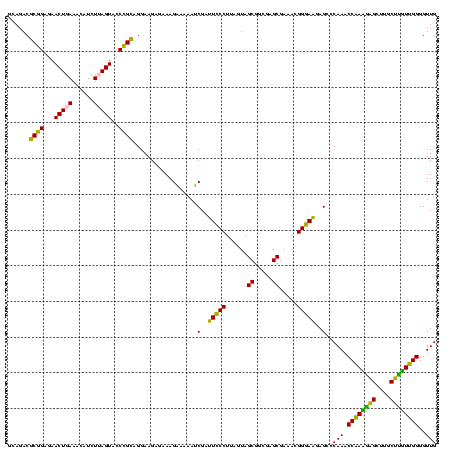
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Mean single sequence MFE | -38.46 |
| Consensus MFE | -35.48 |
| Energy contribution | -32.38 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2720-2840 GAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAA ..(((.....(((((((..(((....)))(((((((......))))))).))(((((((.........)))))))....)).))).((((.(.(((((......))))).))))).))). ( -38.60) >sy_au.0 2275427 120 - 2809422/2720-2840 GAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAA ..(((........((((..(((....)))(((((((......))))))).))(((((((.........)))))))....)).....((((.(.(((((......))))).))))).))). ( -36.70) >sy_sa.0 1670096 120 - 2516575/2720-2840 GAUCCAGAGAUUUCCUAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAA ..(((...(((((.....((((....))))....)))))..(((.(((..(((((((((.........))))))).))..))))))((((.(.(((((......))))).))))).))). ( -38.90) >sp_pn.0 1851953 113 + 2038615/2720-2840 GAUCCAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAA ..(((.....(((((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).)))))..(((((..(((..........))).))))).))). ( -42.70) >sp_ag.0 1905165 113 - 2160267/2720-2840 GAUCCAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAA ..(((.......(((....(((....)))((((--(((...)))-)))).(((((((((.----....))))))).)).)))....(((((..(((..........))).))))).))). ( -35.40) >consensus GAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCA_GAGUUAUGUC_UGUUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAGGCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAA ..(((........((....(((....)))(((((((......))))))).(((((((((.........))))))).)).)).....((((...(((((......)))))..)))).))). (-35.48 = -32.38 + -3.10) # Strand winner: reverse (1.00)

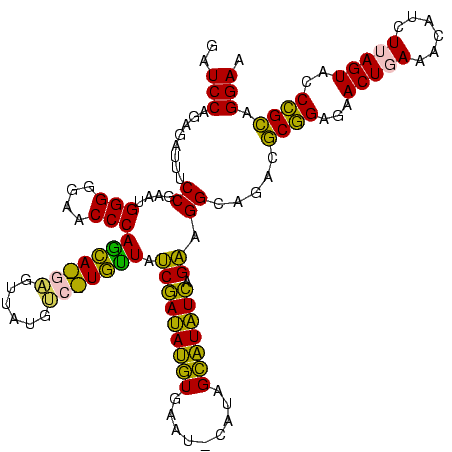
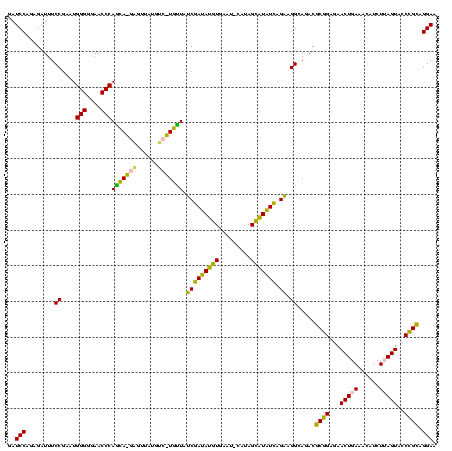
| Location | 2,539,203 – 2,539,323 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.81 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -33.96 |
| Energy contribution | -31.10 |
| Covariance contribution | -2.86 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 120 + 2685015/2760-2880 ACUAACGACGAUAUGCUUUGGGUAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAG ..((((.((((((((((((((((.(((.......)))....)))))))).....((..((((....))))...))...))))))))))))..(((((((.........)))))))..... ( -40.30) >sy_au.0 2275427 120 - 2809422/2760-2880 ACUAACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAG .........((((((((((((((((((.......)))))...))))))......(((..(((....)))(((((((......))))))).))).(((((....))))))))))))..... ( -37.50) >sy_sa.0 1670096 120 - 2516575/2760-2880 ACUAACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCUAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAG ..((((.((((((((((((((((((((.......)))))...)))))))..(((....((((....))))....))).))))))))))))(((((((((.........))))))).)).. ( -38.80) >sp_pn.0 1851953 113 + 2038615/2760-2880 ACAAACGACGAUAUGCCUUGGGUAGCUGUAAGUAAGCGAUGAUCCAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAG ........((.....((((((((.(((.......)))....)))))))).....))...(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).. ( -41.70) >sp_ag.0 1905165 113 - 2160267/2760-2880 ACUAACGACGAAAUGCUUUGGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAG ......((((.....(((((((.(((.((......)))))..))))))).)))).....(((....)))((((--(((...)))-)))).(((((((((.----....))))))).)).. ( -33.70) >consensus ACUAACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCA_GAGUUAUGUC_UGUUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAG .........((....(((((((..(((.......))).....)))))))...)).....(((....)))(((((((......))))))).(((((((((.........))))))).)).. (-33.96 = -31.10 + -2.86) # Strand winner: reverse (1.00)

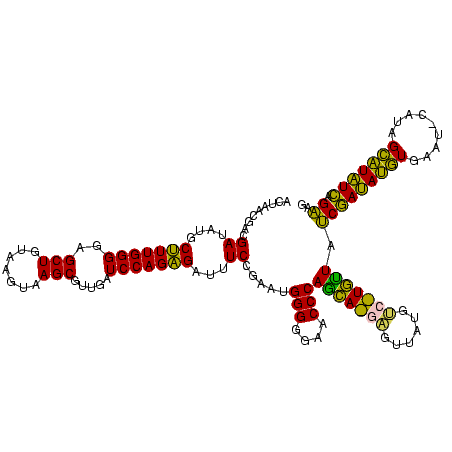
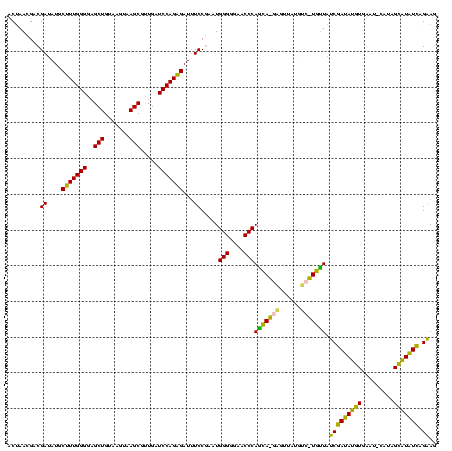
| Location | 2,539,203 – 2,539,300 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 55.69 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -8.82 |
| Energy contribution | -9.20 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.09 |
| Structure conservation index | 0.39 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 97 + 2685015/2960-3080 UAAAAAAGC-UUGAAUUC-AUAAAAUAUAAUCGCUAGUGUUCGAAAGAACACUCACA-AGAUUAAUAACAGGUU----UCUU-------AAAAUUAGAACUACGUUCUAAU--------- .....((((-(((.(((.-....))).(((((...(((((((....)))))))....-.)))))....))))))----)...-------...((((((((...))))))))--------- ( -22.70) >sy_au.0 2275427 117 - 2809422/2960-3080 UAAAUAAGC-UUGAAUUC-AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACA-AGAUUAAUAACGCGUUUAAAUCUUUUUAUAAAAGAACGUAACUUCAUGUUAACGUUUGACUU ....(((((-.((..(((-....))).(((((...(((((((....)))))))....-.)))))....)).)))))...............(((((((((.....))).))))))..... ( -24.40) >sy_sa.0 1670096 100 - 2516575/2960-3080 UUAACAAGCUUUGAAUUCAAAAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCAC--AGAUUAAUAACAUUUU-GGGUUUU-------CAACCGACUUGUUCGUGUUGA---------- ............((((((((((.(...(((((...(((((((....)))))))...--.)))))....).))))-))))))(-------(((((((.....))).)))))---------- ( -26.80) >sp_ag.0 1905165 104 - 2160267/2960-3080 AAUAUCUAUAUCAAAUUCCACGAUCUAGAAAUA-GAUUGUAGAAAGUAACAAGAAAAUAAACCGAAAACGCUGU-GAAUAUU-------UAAUGAGUUUUCUAGUUUUAAAGA------- ....(((........(((.((((((((....))-)))))).)))...................((((((....(-((....)-------))....)))))).........)))------- ( -17.30) >consensus UAAAUAAGC_UUGAAUUC_AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACA_AGAUUAAUAACACGUU_GAAUCUU_______AAAAAAAUAACUUCGUGUUAA__________ ...........................(((((...(((((((....)))))))......)))))........................................................ ( -8.82 = -9.20 + 0.38) # Strand winner: reverse (0.96)

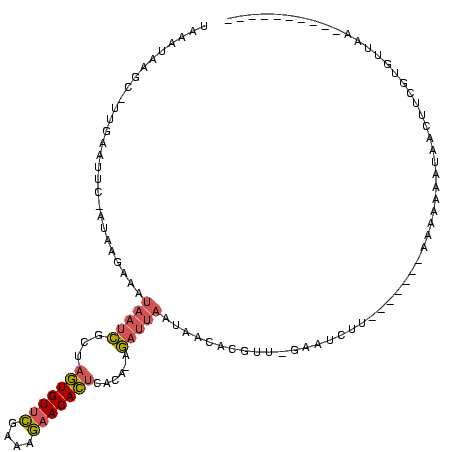
| Location | 2,539,203 – 2,539,308 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.19 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -10.26 |
| Energy contribution | -8.95 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2539203 105 + 2685015/2975-3095 GUGAAU-AAGAGUUUUAAAAAAGC-UUGAAUUC-AUAAAAUAUAAUCGCUAGUGUUCGAAAGAACACUCACA-AGAUUAAUAACAGGUU----UCUU-------AAAAUUAGAACUACGU ((((((-..((((((.....))))-))..))))-))......(((((...(((((((....)))))))....-.)))))......((((----.((.-------......)))))).... ( -24.70) >sy_au.0 2275427 117 - 2809422/2975-3095 GUGAAUAAAGAGUUUUAAAUAAGC-UUGAAUUC-AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACA-AGAUUAAUAACGCGUUUAAAUCUUUUUAUAAAAGAACGUAACUUCAU ((((((...((((((.....))))-))..))))-))..(((.(((((...(((((((....)))))))....-.))))).....((((.....(((((.....)))))))))...))).. ( -27.60) >sy_sa.0 1670096 110 - 2516575/2975-3095 GUGAAUUAGAGUUUUUUAACAAGCUUUGAAUUCAAAAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCAC--AGAUUAAUAACAUUUU-GGGUUUU-------CAACCGACUUGUUCGU .(((((((((((((......)))))))).)))))........(((((...(((((((....)))))))...--.)))))..((((..((-((.....-------...))))..))))... ( -30.10) >sp_ag.0 1905165 111 - 2160267/2975-3095 GUUCAUUGAAAAUUGAAUAUCUAUAUCAAAUUCCACGAUCUAGAAAUA-GAUUGUAGAAAGUAACAAGAAAAUAAACCGAAAACGCUGU-GAAUAUU-------UAAUGAGUUUUCUAGU (((((.(....).)))))............(((.((((((((....))-)))))).)))...................((((((....(-((....)-------))....)))))).... ( -20.20) >consensus GUGAAUUAAAAGUUUUAAAUAAGC_UUGAAUUC_AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACA_AGAUUAAUAACACGUU_GAAUCUU_______AAAAAAAUAACUUCGU ((((.......((((.....))))...........................((((((....))))))))))................................................. (-10.26 = -8.95 + -1.31) # Strand winner: reverse (1.00)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:00:47 2006