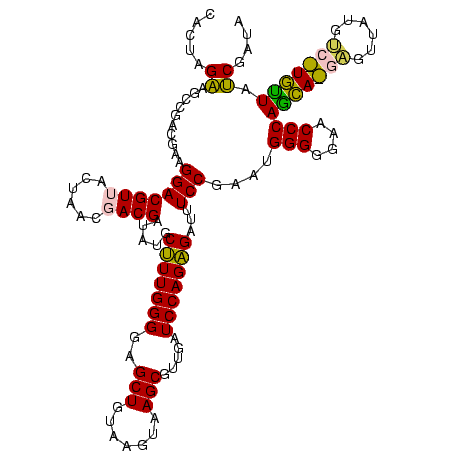
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 973,169 – 973,289 |
| Length | 120 |
| Max. P | 0.999993 |

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.47 |
| Mean single sequence MFE | -35.86 |
| Consensus MFE | -28.20 |
| Energy contribution | -27.50 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/40-160 CACUAGAAGCCGACGAAGGACGUUACUAACGACGAUAUGCUUUGGGUAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACA ........(((......)).)(((..((((.((((((((((((((((.(((.......)))....)))))))).....((..((((....))))...))...))))))))))))..))). ( -36.10) >sy_au.0 694299 120 - 2809422/40-160 CACUAGAAGCCGAUGAAGGACGUUACUAACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUA ..........((((((..(((((.(((.....((.....((((((((((((.......)))))...))))))).....))..((((....)))).....))).)))))...))))))... ( -37.50) >sy_sa.0 745546 120 + 2516575/40-160 CACUAGAAGUCGACGAAGGACGUUACUAACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUA ........(((((....(((((((......)))).....((((((((((((.......)))))...)))))))...)))....(((....)))(((((((......))))))).))))). ( -38.00) >sp_pn.0 16940 117 + 2038615/40-160 CACUAGGAGCCGACGAAGGACGUGACAAACGACGAUAUGCCUUGGGUAGCUGUAAGUAAGCGAUGAUCCAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACA .....((((((......)).(((.....)))........((((((((.(((.......)))....))))))))..))))....(((....)))((((--((.....))-))))....... ( -35.60) >sp_ag.0 350400 117 + 2160267/40-160 CACUAGAAGCCGAAGAAGGACGUGACUAACGACGAAAUGCUUUGGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUA .(((((..(((......)).).((((((..((((.....(((((((.(((.((......)))))..))))))).))))....((((....)))).))--))))....)-))))....... ( -32.10) >consensus CACUAGAAGCCGACGAAGGACGUUACUAACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCA_GAGUUAUGUC_UGUUAUCGAUA .....((..........(((((((......)))).....(((((((..(((.......))).....)))))))...)))....(((....)))(((((((......))))))).)).... (-28.20 = -27.50 + -0.70)


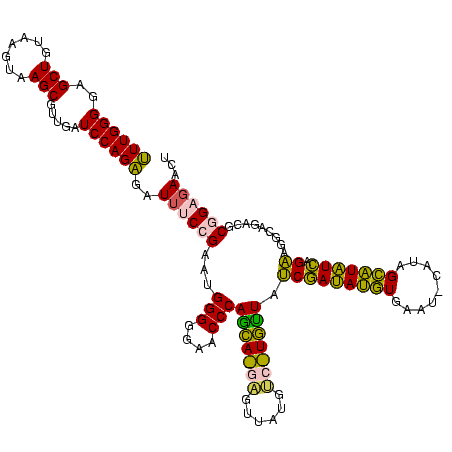
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.47 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -32.34 |
| Energy contribution | -30.48 |
| Covariance contribution | -1.86 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.36 |
| SVM RNA-class probability | 0.992918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/80-200 UUUGGGUAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACU (((((((.(((.......)))....)))))))..((((((..((((....))))...........(((.(((..(((((((((.........))))))).))..)))))).))))))... ( -39.60) >sy_au.0 694299 120 - 2809422/80-200 UUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACU (((((((((((.......)))))...))))))..((((((...(((....)))(((((((......)))))))...(((((((.........)))))))............))))))... ( -37.40) >sy_sa.0 745546 120 + 2516575/80-200 UUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAGACGCGGAGAACU (((((((((((.......)))))...))))))..((((((..((((....))))...........(((.(((..(((((((((.........))))))).))..)))))).))))))... ( -39.80) >sp_pn.0 16940 113 + 2038615/80-200 CUUGGGUAGCUGUAAGUAAGCGAUGAUCCAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAGACGCAGUGAACU (((((((.(((.......)))....)))))))..(((((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).)))))............ ( -41.80) >sp_ag.0 350400 113 + 2160267/80-200 UUUGGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACU ((((((.(((.((......)))))..))))))....(((....(((....)))((((--(((...)))-)))).(((((((((.----....))))))).)).))).............. ( -31.50) >consensus UUUGGGGAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCA_GAGUUAUGUC_UGUUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAGGCAGACGCGGAGAACU ((((((..(((.......))).....))))))..((((((...(((....)))(((((((......))))))).(((((((((.........))))))).)).........))))))... (-32.34 = -30.48 + -1.86)


| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -34.46 |
| Consensus MFE | -31.36 |
| Energy contribution | -27.78 |
| Covariance contribution | -3.58 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/120-240 AAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGA ..((((....))))...(((.....(((.(((..(((((((((.........))))))).))..))))))((((.(.(((((......))))).))))).................))). ( -35.40) >sy_au.0 694299 120 - 2809422/120-240 AAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGA ..((((....))))...((((((...((.(((..(((((((((.........))))))).))..)))(..((((.(.(((((......))))).)))))..)........)).)))))). ( -34.60) >sy_sa.0 745546 120 + 2516575/120-240 AAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGA ..((((....))))...(((.....(((.(((..(((((((((.........))))))).))..))))))((((.(.(((((......))))).))))).................))). ( -36.80) >sp_pn.0 16940 113 + 2038615/120-240 AAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGA ...(((....)))((((--((.....))-)))).(((((((((.----....))))))).))........(((((..(((..........))).)))))...........((....)).. ( -35.60) >sp_ag.0 350400 113 + 2160267/120-240 AAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGA ...(((....)))((((--(((...)))-)))).(((((((((.----....))))))).))........(((((..(((..........))).)))))...........((....)).. ( -29.90) >consensus AAUGGGGGAACCCAGCA_GAGUUAUGUC_UGUUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAGGCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGA ...(((....)))(((((((......))))))).(((((((((.........))))))).))........((((...(((((......)))))..))))...........((....)).. (-31.36 = -27.78 + -3.58)


| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -24.32 |
| Consensus MFE | -21.30 |
| Energy contribution | -17.76 |
| Covariance contribution | -3.54 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.611938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/120-240 UCGAUUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUCUGCCUUCUGACAAGCUAUGAAUUCACUUGUCGAUAACACGACAUAACUCGUGCUGGGUUCCCCCAUU ..((..(.((((......((....))....)))).)..))........(((...((((..(((((((...((....)))))))))....(((((......)))))..))))...)))... ( -25.10) >sy_au.0 694299 120 - 2809422/120-240 UCGAAUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUGUGCCUUCUGAUAUGCUAUGUAUUCACAUAUCGAUAACAUGACAUAACUCAUGCUGGGUUUCCCCAUU ..(((...((((......((....))....))))...)))........(((...((((..(((((((...........)))))))....(((((......)))))..))))...)))... ( -22.80) >sy_sa.0 745546 120 + 2516575/120-240 UCGAUUUUUCUUUCUCUUCCUGCGGGUACUAAGAUGUUUCAGUUCUCCGCGUCUGCCUUCAGAUAUGCUAUGUAUUCACAUAUCGAUAACACGACAUAACUCGUGCUGGGUUUCCCCAUU ((((.................(((((.(((..((....))))).).))))(((((....)))))....(((((....)))))))))...(((((......))))).((((....)))).. ( -24.90) >sp_pn.0 16940 113 + 2038615/120-240 UCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUCCUCACAUCCUUAA----CAGAUGUGGGUAACA-GGUAUUACC--UGUUGGGUUCCCCCAUU ..((....))...........((((..(((..((....)))))...)))).........((((((((.....----..)))))))).((((-((.....))--))))(((....)))... ( -27.70) >sp_ag.0 350400 113 + 2160267/120-240 UCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAA----CAGAUAUGGAUACUA-GUCAUUAAC--UAGUGGGUUCCCCCAUU ..((....))...........((((..(((..((....)))))...))))........((.((((((.....----..)))))))).((((-((.....))--))))(((....)))... ( -21.10) >consensus UCGAUUUUUCUUUCUCUUCCUGCGGGUACUAAGAUGUUUCAGUUCUCCGCGUCUGCCUUCUGAUAUGCUAUG_AUUCACAUAUCGAUAACA_GACAUAACUC_UGCUGGGUUCCCCCAUU ..((....))..........(((((..(((..((....)))))...))))).......((.((((((...........))))))))...(((((......))))).((((....)))).. (-21.30 = -17.76 + -3.54) # Strand winner: forward (1.00)

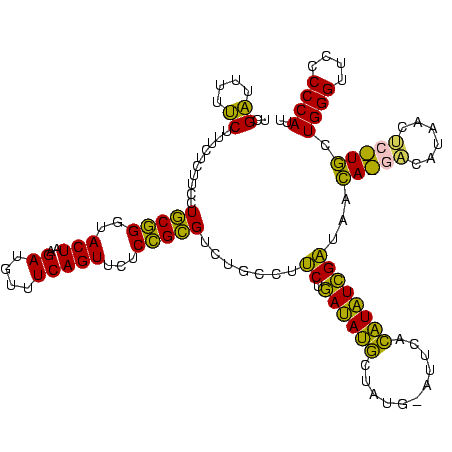

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -34.36 |
| Consensus MFE | -27.60 |
| Energy contribution | -24.52 |
| Covariance contribution | -3.08 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/200-320 GAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACG ......((((..(((.(((.((((..((........))..))))))).)))(((((.........(((.....)))...((((((((....)))))))).)))))))))........... ( -30.90) >sy_au.0 694299 120 - 2809422/200-320 GAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACG ...........(((((((..((((..(((......)))..)))).......((.....))....)))).(((((((((.((((((((....))))))))..)))..))...)))).))). ( -33.40) >sy_sa.0 745546 120 + 2516575/200-320 GAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACG ...........(((((((((((((..((........))..)).))))....((.....))....)))).(((((((((.((((((((....))))))))..)))..))...)))).))). ( -31.80) >sp_pn.0 16940 119 + 2038615/200-320 GAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUG ...((((....((.((((((((....((..((....))..)).)))..))))).))..........((((....((((.((((((((....))))))))..))))....)-))).)))). ( -38.10) >sp_ag.0 350400 119 + 2160267/200-320 GAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUA .....(((...((.((((((((....((..((....))..)).)))..))))).))..........((((....((((.((((((((....))))))))..))))....)-))))))... ( -37.60) >consensus GAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAAGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACG ......(((((.((.((((...........((....))..(((((......((.....)).....)))))....))...((((((((....)))))))))).)))))))........... (-27.60 = -24.52 + -3.08)

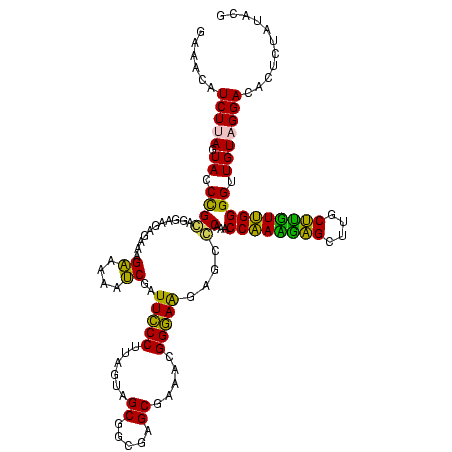
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -31.44 |
| Consensus MFE | -22.32 |
| Energy contribution | -19.44 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/200-320 CGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUC .((((.(((((....((....((((((((....))))))))...(((.....)))))..))))).(((......((((((..((....)).)))))).....)))))))........... ( -30.40) >sy_au.0 694299 120 - 2809422/200-320 CGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUC .((((.(((((....((....((((((((....))))))))...(((.....)))))..))))).(((.....(((((((..((....)).)))))))....)))))))........... ( -32.60) >sy_sa.0 745546 120 + 2516575/200-320 CGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUCUUCCUGCGGGUACUAAGAUGUUUC .((((.(((((....((....(((((.((....)).)))))...(((.....)))))..))))).((((.....((((((..((....)).))))))....))))))))........... ( -32.50) >sp_pn.0 16940 119 + 2038615/200-320 CACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUC .((((((((-(.....(((..((((((((....)))))))).))).....))))......(((.((.......(((((((......)))))))........)))))......)))))... ( -30.56) >sp_ag.0 350400 119 + 2160267/200-320 UAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUC .((((((((-(.....(((..((((((((....)))))))).))).....))))......(((.((.......(((((((......)))))))........)))))......)))))... ( -31.16) >consensus CGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAUUUUUCUUUCUCUUCCUGCGGGUACUAAGAUGUUUC ..........(((........((((((((....)))))))).((((.....((((.....((.....))......))))...((....))...........)))).......)))..... (-22.32 = -19.44 + -2.88) # Strand winner: forward (0.99)

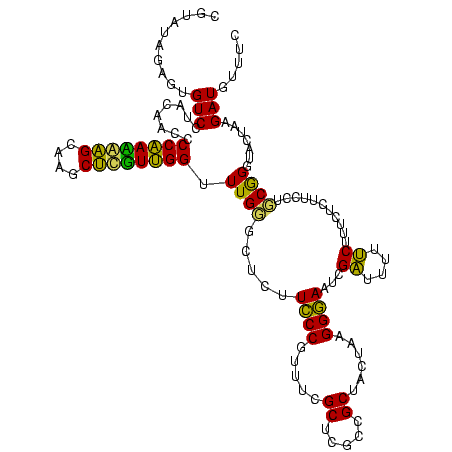

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -20.56 |
| Energy contribution | -19.36 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/240-360 ACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAA .......(((((...)))))......(((((..(((....((....(((((....((....((((((((....))))))))...(((.....)))))..)))))..)).)))..))))). ( -28.00) >sy_au.0 694299 120 - 2809422/240-360 UCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAA ...((((.((((((.........))))))(((.(((....((....(((((....((....((((((((....))))))))...(((.....)))))..)))))..)).))).))))))) ( -31.50) >sy_sa.0 745546 120 + 2516575/240-360 UCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAA ...(((((((((...))))).....................((...(((((....((....(((((.((....)).)))))...(((.....)))))..)))))...))......)))). ( -24.50) >sp_pn.0 16940 113 + 2038615/240-360 UCUUCCCAAAUCAUUCUUCUAUAAUCUU------UGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAA ............................------.((((....((.(((-(.....(((..((((((((....)))))))).))).....)))).))...))))(((........))).. ( -27.20) >sp_ag.0 350400 113 + 2160267/240-360 CCUUCCCAGGUAAUUCUUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAA .......(((..(((..((......)).------.)))))).....(((-(.....(((..((((((((....)))))))).))).....))))..........(((........))).. ( -25.40) >consensus UCUUUCCAGAUGAUUCGUCUAAUAUCUUC_UUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAA .....(((((..........................((((......))))...........((((((((....))))))))))))).....((((.....((.....))......)))). (-20.56 = -19.36 + -1.20) # Strand winner: forward (1.00)

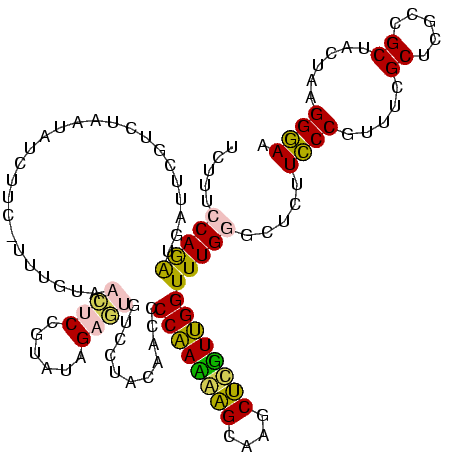
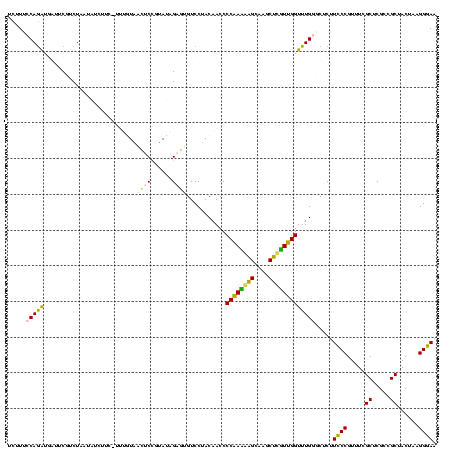
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -27.80 |
| Energy contribution | -22.46 |
| Covariance contribution | -5.34 |
| Combinations/Pair | 1.82 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/280-400 CAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAU (((((((....)))))))...(((((....(((((....))))))))))....(((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...))))))) ( -23.90) >sy_au.0 694299 120 - 2809422/280-400 CAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUG (((((((....)))))))...(((((....(((((....))))))))))....(((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))) ( -35.00) >sy_sa.0 745546 120 + 2516575/280-400 CAACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGU (((((((....)))))))...(((((....(((((....))))))))))....(((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))) ( -35.50) >sp_pn.0 16940 113 + 2038615/280-400 CGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCA------AAGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGUCU ....((((((((((....))((((((....-)))))))))))))).------.(((((((....(..((((((....))))))..)...(((.((....))))).........))))))) ( -27.90) >sp_ag.0 350400 113 + 2160267/280-400 CGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUA------AUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAG (((((((....)))))))..((((((....-)))))).........------((((((.((((.(..((((((....))))))..)...(((.((....)))))...)))).)))))).. ( -29.30) >consensus CAAAGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAA_GAACAUAAUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAU (((((((....)))))))....(((((......)))))...............(((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))) (-27.80 = -22.46 + -5.34)

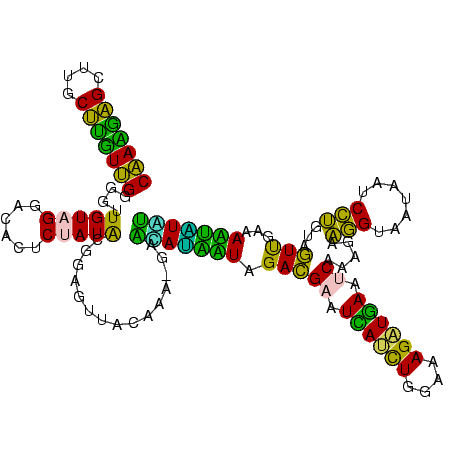
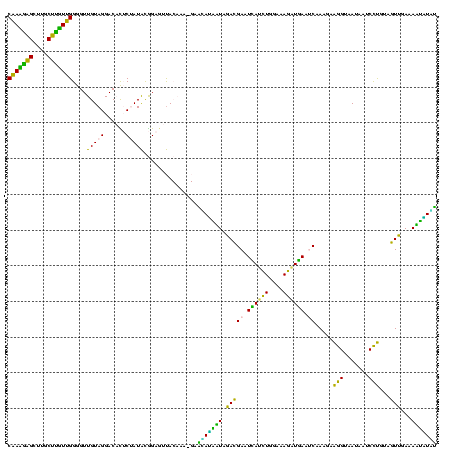
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -20.21 |
| Consensus MFE | -18.54 |
| Energy contribution | -14.70 |
| Covariance contribution | -3.84 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/280-400 AUAUAUUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUG ................((((.(((.........(((.(((.((....)).))).)))......))).))))(((((((((......))))....)))))...(((((((....))))))) ( -16.96) >sy_au.0 694299 120 - 2809422/280-400 CAACAUUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUG ..(((((...(((...(((.......))).....((.((((((....)))))).))))).)))))......(((((((((......))))....)))))...(((((((....))))))) ( -24.80) >sy_sa.0 745546 120 + 2516575/280-400 ACAAACUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUG ..........((((((((((..((...((....(((.((((((....)))))).)))...))...))..)))))))((((......))))))).........((((.((....)).)))) ( -21.20) >sp_pn.0 16940 113 + 2038615/280-400 AGACUAUUUUAAAUACGAGGCUAUUACUCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUAUAAUCUU------UGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCG .((((...........(((.......)))..(((((.........)))))..................------..)))).........-............(((((((....))))))) ( -16.80) >sp_ag.0 350400 113 + 2160267/280-400 CUAUCAUUUUAAAUACGAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCG .................(((((((..........(..((((((....))))))..).........(((------.........))))))-).))).......(((((((....))))))) ( -21.30) >consensus AUACAAUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUAUCUUC_UUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUCGUUG (((((((.........(((.......))).....((.((((((....)))))).))....)))))))...................................(((((((....))))))) (-18.54 = -14.70 + -3.84) # Strand winner: forward (1.00)

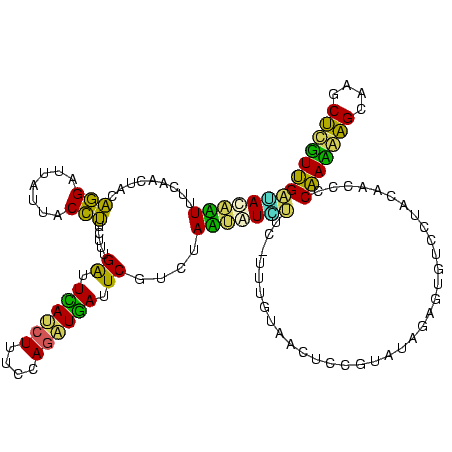
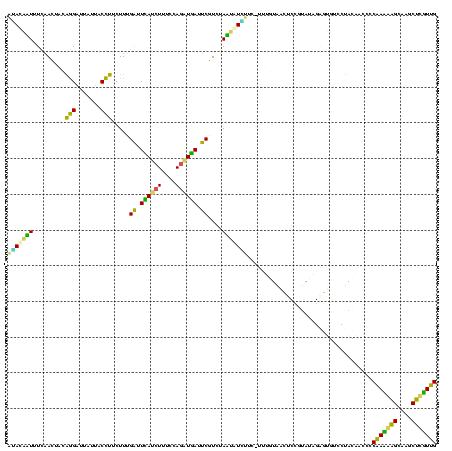
| Location | 973,169 – 973,288 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.31 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -19.14 |
| Energy contribution | -15.40 |
| Covariance contribution | -3.74 |
| Combinations/Pair | 1.92 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 119 + 2685015/320-440 GAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAU-UCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGA ...((((...((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...)))))))-))).......(((..(((..........))).))))))) ( -23.80) >sy_au.0 694299 119 - 2809422/320-440 GAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUG-UCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGA ...((((...((((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))-))).......(((..(((..........))).))))))) ( -33.60) >sy_sa.0 745546 119 + 2516575/320-440 GAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGU-UCACUCUUGAGUGGAUCCUGAGUACGACGGAACACGAGA ((((.......(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))-))))))....(((..(((..........))).))).... ( -32.81) >sp_pn.0 16940 113 + 2038615/320-440 GACUCA------AAGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGUCU-UUGUACUUAGCAGUAUCCUGAGUACGGCGGGACACGUGA ..((((------((((((((....(..((((((....))))))..)...(((.((....))))).........)))))))-))((((((((.......))))))))...)))........ ( -26.30) >sp_ag.0 350400 114 + 2160267/320-440 GGAUUA------AUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGUACGGCGAGACACGCGA .((((.------((((((.((((.(..((((((....))))))..)...(((.((....)))))...)))).)))))).))))...(((((.......)))))....(((.....))).. ( -23.10) >consensus GAGUUACAAA_GAACAUAAUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAU_UCACUCUUGAGUGGAUCCUGAGUACGACGGAACACGUGA .............(((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))...........................(((.....))).. (-19.14 = -15.40 + -3.74)

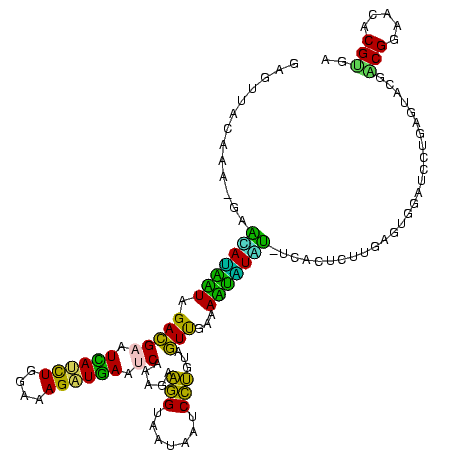
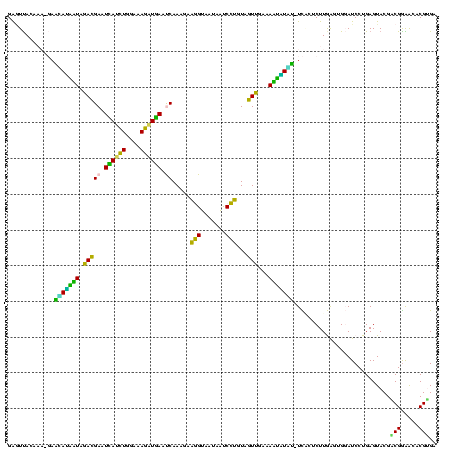
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -31.68 |
| Energy contribution | -30.56 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/440-560 AAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAGAGAACU ..(((((((((....((((((......))))))..((((.........))))..))))).....(((....((....))....)))........((((....)))).).)))........ ( -33.80) >sy_au.0 694299 120 - 2809422/440-560 AAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACC ..(((((((((....((((((......))))))..((((.........))))..))))).((..(((....((....))....))).....)).((((....)))).).)))........ ( -34.80) >sy_sa.0 745546 120 + 2516575/440-560 AAUUCCGUCGGAAUCUGGGAGGACCAUCUCCCAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAUAGAACU ..(((((((((....((((((......))))))..((((.........))))..))))).....(((....((....))....)))........((((....)))).).)))........ ( -36.10) >sp_pn.0 16940 120 + 2038615/440-560 AAUCCCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCCCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACC ..(((....)))...((((((......))))))...(((..((((((((.....(((...(((.(((....((....))....)))......)))..)))..))))))))...))).... ( -36.90) >sp_ag.0 350400 120 + 2160267/440-560 AAUCUCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACC ..((.((((((....((((((......))))))........((((....)))).))))).).))(((....((....))....))).....((.((((....)))).))........... ( -35.40) >consensus AAUUCCGUCGGAAUCUGGGAGGACCAUCUCCCAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACC ..((((..(((....((((((......))))))...........((.(((........))).)).......)))...))))..(((.....((.((((....)))).))........))) (-31.68 = -30.56 + -1.12)

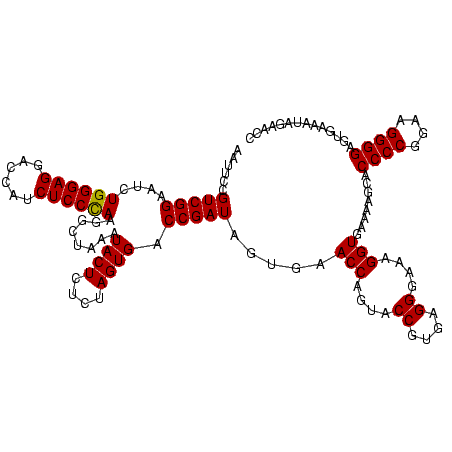
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.67 |
| Mean single sequence MFE | -35.99 |
| Consensus MFE | -34.62 |
| Energy contribution | -33.58 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/520-640 AAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAGAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACG ..............((((....))))..........((((((...(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))))))))... ( -35.80) >sy_au.0 694299 120 - 2809422/520-640 AAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACG ..((((.....((.((((....)))).))........))))....(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))......... ( -36.52) >sy_sa.0 745546 120 + 2516575/520-640 AAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAUAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACG ..............((((....))))..........((((((...(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))))))))... ( -35.80) >sp_pn.0 16940 120 + 2038615/520-640 AAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACG ..((((.....((.((((....)))).))........))))....(((.((..((((((((.((((..(((((....)))))..)))).))....)))))).....)))))......... ( -35.92) >sp_ag.0 350400 120 + 2160267/520-640 AAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACG ..((((.....((.((((....)))).))........))))....(((.((..((((((((.((((..(((((....)))))..)))).))....)))))).....)))))......... ( -35.92) >consensus AAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACG ....((((...((.((((....))))....................((.((..(((((((..((((..(((((....)))))..))))......))))))).....))))))...)))). (-34.62 = -33.58 + -1.04)


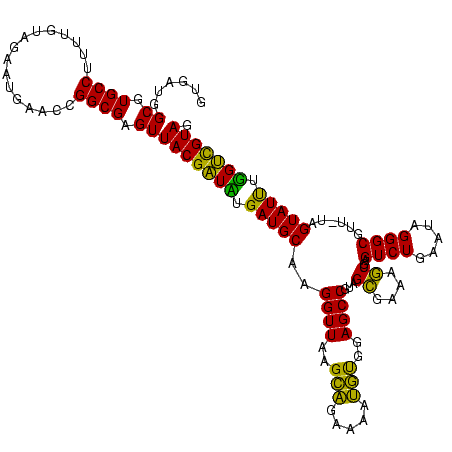
| Location | 973,169 – 973,288 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.72 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -34.85 |
| Energy contribution | -31.89 |
| Covariance contribution | -2.96 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.33 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.820187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 119 + 2685015/600-720 GUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCUGAUGCAAGGUUAAGCAGGAAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU-GAGUAUUUGGUCGUAG ......((.((((................)))).))(((((((.(((((..((((..(((.....)))..))))...((....))..((((.....))))...-..))))).))))))). ( -33.09) >sy_au.0 694299 119 - 2809422/600-720 GUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUUUGAUGCAAGGUUAAGCAGUAAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU-UAGUAUUUGGUCGUAG ......((.((((................)))).))((((((..(((((..((((..(((.....)))..))))...((....))..((((.....))))...-..)))))..)))))). ( -31.79) >sy_sa.0 745546 119 + 2516575/600-720 GUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUUUGAUGCAAGGUUAAGCAGUGAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU-GAGUAUUUGGUCGUAG ......((.((((................)))).))((((((..(((((..((((..(((.....)))..))))...((....))..((((.....))))...-..)))))..)))))). ( -31.79) >sp_pn.0 16940 120 + 2038615/600-720 GUGAGAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGUUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUGAAUAGGGCACCUUAGUAUCAUGACGUAG ((....)).((((................))))...((((((((((((((((((............(((....))).((....))..((((.....))))))))).))))))))))))). ( -38.09) >sp_ag.0 350400 120 + 2160267/600-720 GUGAGAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUUAAUAGGGCGUCAUAGUAUCAUGUUGUAG ((....)).((((................))))...(((((((((((((..((((..(((.....)))..))))...((....))((((((.....)))).))...))))))))))))). ( -36.89) >consensus GUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUAUGAUGCAAGGUUAAGCAGAAAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU_UAGUAUUUGGUCGUAG ......((.((((................)))).))(((((((.(((((..((((..(((.....)))..))))...((....))..((((.....))))......))))).))))))). (-34.85 = -31.89 + -2.96)

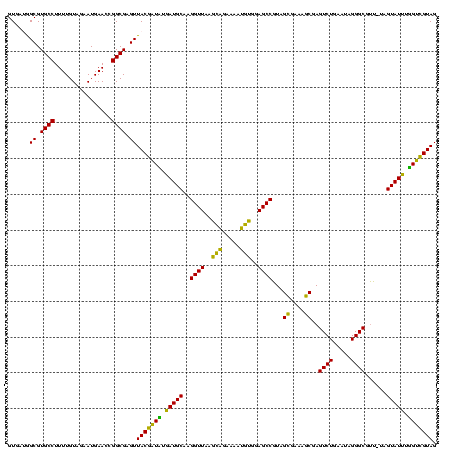
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -42.78 |
| Consensus MFE | -46.44 |
| Energy contribution | -42.04 |
| Covariance contribution | -4.40 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.19 |
| Structure conservation index | 1.09 |
| SVM decision value | 5.78 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/720-840 ACCCGAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAA (((.(....).)))...(((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))))))))))).((((...)))).. ( -41.90) >sy_au.0 694299 120 - 2809422/720-840 ACCCGAAACCAGGUGAUCUACCCUUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAGAAAUUCCAA (((.(....).)))...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).((((...)))).. ( -42.20) >sy_sa.0 745546 120 + 2516575/720-840 ACCCGAAACCAGGUGAUCUACCCAUGACCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGUAGCGGAGAAAUUCCAA (((.(....).)))...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).((((...)))).. ( -42.10) >sp_pn.0 16940 120 + 2038615/720-840 ACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAGACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAA .................(((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))))).((((...)))).. ( -44.50) >sp_ag.0 350400 120 + 2160267/720-840 ACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAA .................(((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))))).((((...)))).. ( -43.20) >consensus ACCCGAAACCAGGUGAUCUACCCAUGAGCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAAUUGUGGGUAGCGGAGAAAUUCCAA .................(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).((((...)))).. (-46.44 = -42.04 + -4.40)

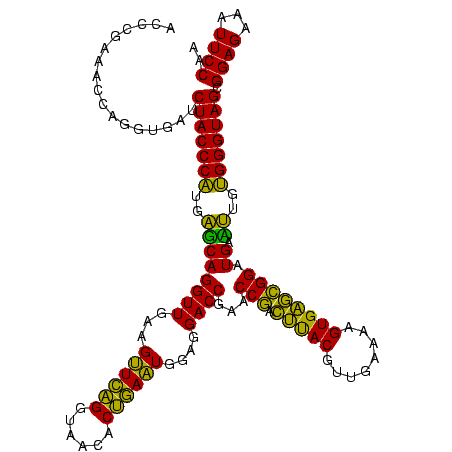
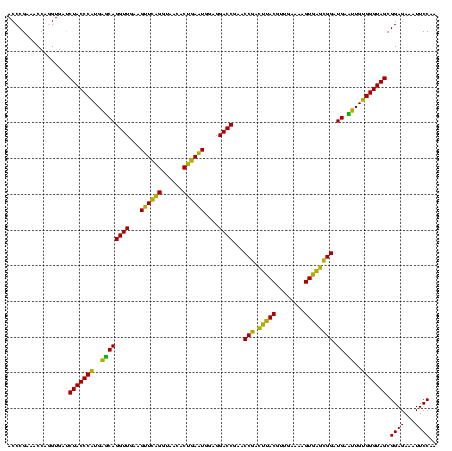
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.42 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -36.21 |
| Energy contribution | -33.49 |
| Covariance contribution | -2.72 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/920-1040 UGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCA ((((((((.((((....))))((((....)))).)))))))).(((((.((................)).)))))..(((((((((........)))))))))....(((......))). ( -40.39) >sy_au.0 694299 120 - 2809422/920-1040 UGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCA ((((((((.((((....))))((((....)))).)))))))).(((((.((................)).)))))..(((((((((........)))))))))....(((......))). ( -40.39) >sy_sa.0 745546 120 + 2516575/920-1040 UGUUUGGACGAGGGGCCCUUAUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUCAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCA ((((((((...(((((((.....))))).))...)))))))).(((((.((................)).)))))..(((((((((........)))))))))....(((......))). ( -38.99) >sp_pn.0 16940 120 + 2038615/920-1040 UGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAAUGAAUUAUGGUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCA ....(((((...(..(((......((((((..((((.((....((((((...((((........))))))((((.....)))))))).)).))))..)))))).....)))..).))))) ( -36.50) >sp_ag.0 350400 120 + 2160267/920-1040 UGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAGAUAUAAUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCA ....(((((...(..(((......((((((..((((.((....((((..........(((.......)))((((.....)))))))).)).))))..)))))).....)))..).))))) ( -35.40) >consensus UGUUUGGACGAGGGGCCCAUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUGAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCA ((((((((...(((((((.....))))).))...)))))))).(((((((.................)))).)))..((((((((((......))))))))))....(((......))). (-36.21 = -33.49 + -2.72)



| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.75 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -27.08 |
| Energy contribution | -25.80 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/960-1080 CAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAA .............(((.(((((.((..(((((((((.(((((((((........)))))))))....(((......))).)))..((......))))))))..))...))))).)))... ( -33.10) >sy_au.0 694299 120 - 2809422/960-1080 CAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAA .............(((.(((((.((..(((((((((.(((((((((........)))))))))....(((......))).)))..((......))))))))..))...))))).)))... ( -33.10) >sy_sa.0 745546 120 + 2516575/960-1080 CAAACUCCGAAUGCCAAUCAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAA .....(((((.......)).((((((((((((((((.(((((((((........)))))))))....(((......))).)))..((......)))))))).......)))))))))).. ( -33.71) >sp_pn.0 16940 120 + 2038615/960-1080 UAAACUCCGAAUGCCAAUGAAUUAUGGUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAAUUGUUAAGUGGAAA .....((((..((.((((..(((.(((..(((.....(((((((..(......)..))))))).....(....).)))..((((.........))))))).))).)))).))..)))).. ( -30.50) >sp_ag.0 350400 120 + 2160267/960-1080 UAAACUCCGAAUGCCAACGAGAUAUAAUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAA .............(((..((.(((((...(((.....(((((((..(......)..))))))).....(....).)))..((((.........))))......))))).))...)))... ( -27.60) >consensus CAAACUCCGAAUGCCAAUGAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAA .....((((..((.((.............((..(((.((((((((((......))))))))))....(((......))).)))..))......(....)........)).))..)))).. (-27.08 = -25.80 + -1.28)

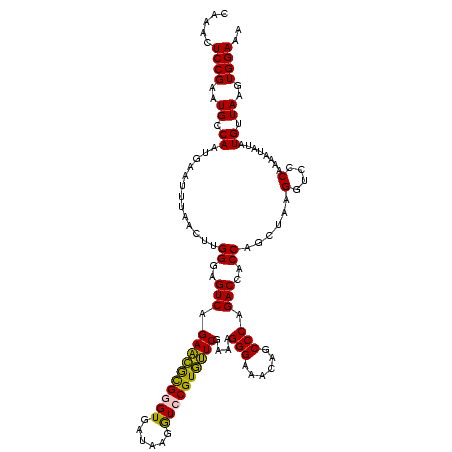
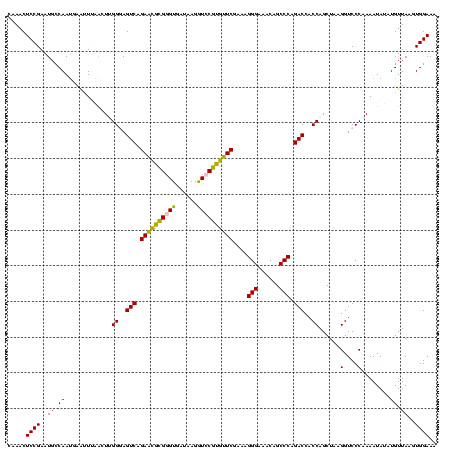
| Location | 973,169 – 973,287 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.98 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -29.34 |
| Energy contribution | -28.38 |
| Covariance contribution | -0.96 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 118 + 2685015/1080-1200 UG-UUUAGCCCCGGUACAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUG-AUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCU ..-....((....))........((((...((((...(((((((.((((......))..........((-.(((((.......))))).)))))))))))....)))).))))....... ( -31.70) >sy_au.0 694299 118 - 2809422/1080-1200 UG-UUUAGCCCCGGUACAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUG-AUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCU ..-....((....))........((((...((((...(((((((.((((......))..........((-.(((((.......))))).)))))))))))....)))).))))....... ( -31.70) >sy_sa.0 745546 118 + 2516575/1080-1200 UG-UUUAGCCCCGGUACAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUG-AUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCU ..-....((....))........((((...((((...(((((((.((((......))..........((-.(((((.......))))).)))))))))))....)))).))))....... ( -31.70) >sp_pn.0 16940 120 + 2038615/1080-1200 UGUUUUAGCCCCGGUACAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCU .......((.((((.......)))).)).(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..))))........ ( -32.10) >sp_ag.0 350400 120 + 2160267/1080-1200 UGUUUUAGCCCCGGUACAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCU .......((.((((.......)))).)).(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..))))........ ( -32.10) >consensus UG_UUUAGCCCCGGUACAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUG_AUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCU .......((.((((.......)))).)).((((((..(((((((.((((......))..............(((((.......)))))...)))))))))..))....))))........ (-29.34 = -28.38 + -0.96) # Strand winner: forward (0.97)

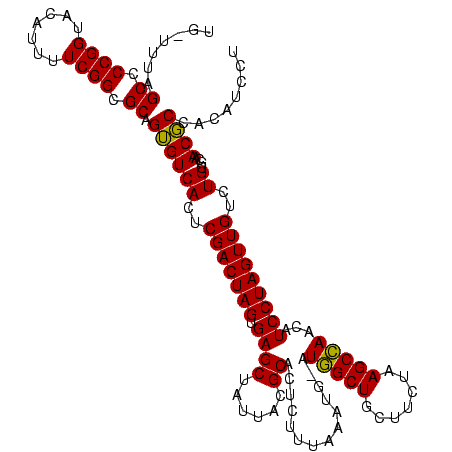
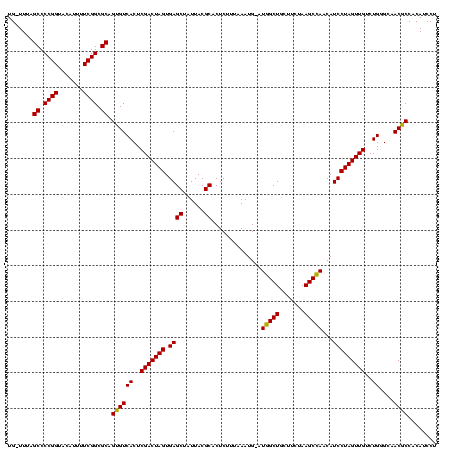
| Location | 973,169 – 973,286 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -33.04 |
| Energy contribution | -31.96 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 117 + 2685015/1120-1240 AAGCAGCCAU-CAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUGUACCGGGGCUAAA-CAUAUUACCGAAGCUGUGGAUUGUCCGUA-GGACAAUGGUAG ..((((...(-((((..(..((((.((....)).))))..)..)))))..))))(((.........(((.((((...-...........)))).)))(((((((...-)))))))))).. ( -35.04) >sy_au.0 694299 117 - 2809422/1120-1240 AAGCAGCCAU-CAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUGUACCGGGGCUAAA-CAUAUUACCGAAGCUGUGGAUUGUCCUUU-GGACAAUGGUAG ..((((...(-((((..(..((((.((....)).))))..)..)))))..))))(((.........(((.((((...-...........)))).)))(((((((...-)))))))))).. ( -35.04) >sy_sa.0 745546 118 + 2516575/1120-1240 AAGCAGCCAU-CAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUGUACCGGGGCUAAA-CAUAUUACCGAAGCUGUGGAUUGUCCGUAAGGACAAUGGUAG ..((((...(-((((..(..((((.((....)).))))..)..)))))..))))(((.........(((.((((...-...........)))).)))(((((((....)))))))))).. ( -39.34) >sp_pn.0 16940 118 + 2038615/1120-1240 AAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAAUUUACCGAAGCUGUGGAUACCUUUAUAGG--UAUGGUAG ..((((....((((((.(..((((.((....)).))))..).))))))..))))(((.........(((.((((...............)))).)))((((((....)))--)))))).. ( -33.96) >sp_ag.0 350400 120 + 2160267/1120-1240 AAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAUAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAG ..((((....((((((.(..((((.((....)).))))..).))))))..))))(((.........(((.((((...............)))).)))(((((((....)))))))))).. ( -36.76) >consensus AAGCAGCCAU_CAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUGUACCGGGGCUAAA_CAUAUUACCGAAGCUGUGGAUUGUCCGUAAGGACAAUGGUAG ..((((.....(((((.(..((((.((....)).))))..).)))))...))))(((.........(((.((((...............)))).)))(((((((....)))))))))).. (-33.04 = -31.96 + -1.08)

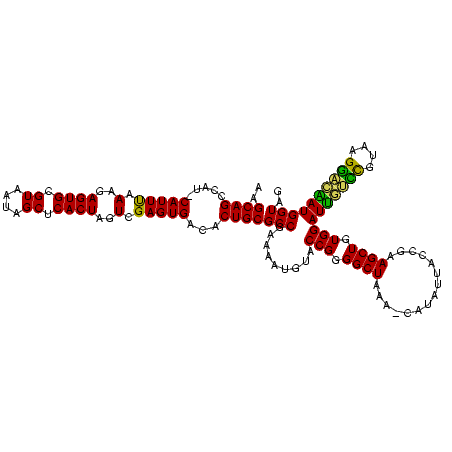
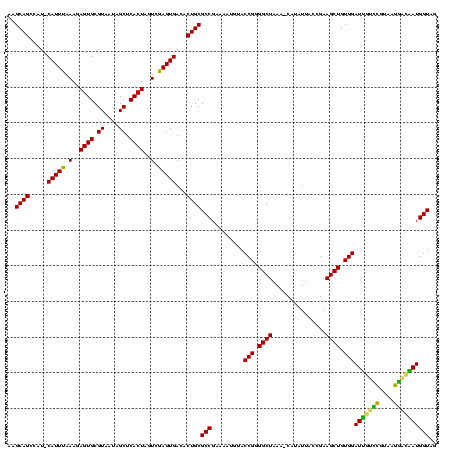
| Location | 973,169 – 973,288 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -40.60 |
| Energy contribution | -37.92 |
| Covariance contribution | -2.68 |
| Combinations/Pair | 1.48 |
| Mean z-score | -3.50 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.88 |
| SVM RNA-class probability | 0.999682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 119 + 2685015/1200-1320 UAUUACCGAAGCUGUGGAUUGUCCGUA-GGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCG ...((((..........(((((((...-)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))))))......... ( -38.90) >sy_au.0 694299 119 - 2809422/1200-1320 UAUUACCGAAGCUGUGGAUUGUCCUUU-GGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCG ...((((..........(((((((...-)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))))))......... ( -37.90) >sy_sa.0 745546 120 + 2516575/1200-1320 UAUUACCGAAGCUGUGGAUUGUCCGUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCG ...((((..........(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))))))......... ( -43.20) >sp_pn.0 16940 118 + 2038615/1200-1320 AUUUACCGAAGCUGUGGAUACCUUUAUAGG--UAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCG ...((((..........((((((....)))--)))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))))))......... ( -39.60) >sp_ag.0 350400 120 + 2160267/1200-1320 UAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCG ...((((..........(((((((....)))))))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))))))......... ( -43.90) >consensus UAUUACCGAAGCUGUGGAUUGUCCGUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCG ...((((..........(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))))))......... (-40.60 = -37.92 + -2.68)

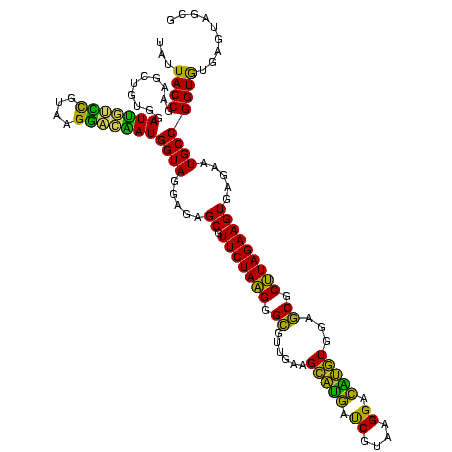
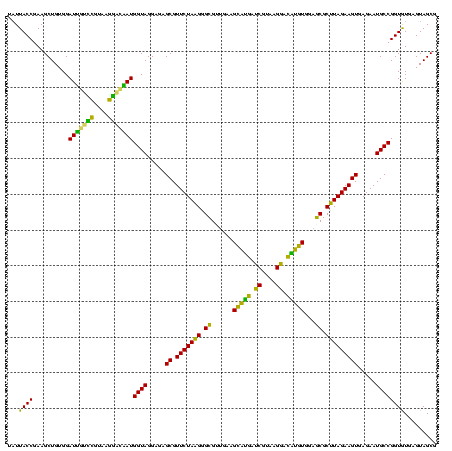
| Location | 973,169 – 973,288 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -19.78 |
| Energy contribution | -17.86 |
| Covariance contribution | -1.92 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 119 + 2685015/1200-1320 CGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCC-UACGGACAAUCCACAGCUUCGGUAAUA ..........(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((((-...)))))))....))).))))).... ( -22.70) >sy_au.0 694299 119 - 2809422/1200-1320 CGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCC-AAAGGACAAUCCACAGCUUCGGUAAUA ..........(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((((-...)))))))....))).))))).... ( -22.70) >sy_sa.0 745546 120 + 2516575/1200-1320 CGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUACGGACAAUCCACAGCUUCGGUAAUA ..........(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((((....)))))))....))).))))).... ( -24.20) >sp_pn.0 16940 118 + 2038615/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCACUCCUCACGGUAUACCUUCAUCACACAUAGAACGCUCUCCUACCAUA--CCUAUAAAGGUAUCCACAGCUUCGGUAAAU .........(((((((..............((((((.............((....)).............)))))).........(((--(((....)))))).....))..)))))... ( -21.07) >sp_ag.0 350400 120 + 2160267/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUGACACUUUUGUGUCAUCCACAGCUUCGGUAAUA .........((((((.........((((((((((....)))).......((....))..........)))))).(((........(((((((....)))))))....))).))))))... ( -25.70) >consensus CGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUAAGGACAAUCCACAGCUUCGGUAAUA ..........(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((((....)))))))....))).))))).... (-19.78 = -17.86 + -1.92) # Strand winner: forward (1.00)

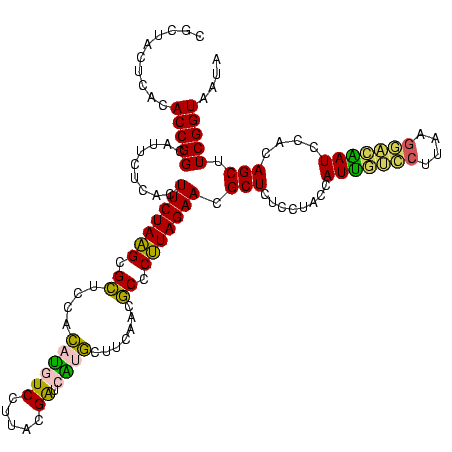
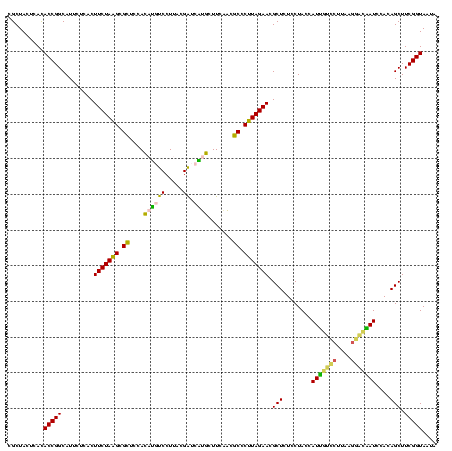
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -37.42 |
| Energy contribution | -36.58 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1240-1360 GAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUU ....(((((((..(((((((...(((((.((....)).)))))..))))))).......)))))))(((((.((....(....).((((.......)))))))))))............. ( -39.90) >sy_au.0 694299 120 - 2809422/1240-1360 GAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUU ....(((((((..(((((((...(((((.((....)).)))))..))))))).......)))))))(((((.((....(....).((((.......)))))))))))............. ( -38.90) >sy_sa.0 745546 120 + 2516575/1240-1360 GAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUU ....(((((((..(((((((...(((((.((....)).)))))..))))))).......)))))))(((((.((....(....).((((.......)))))))))))............. ( -39.90) >sp_pn.0 16940 120 + 2038615/1240-1360 GAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUU ....((.((((((((((......(((((.((....)).)))))...))))))))))))......(((......(((.(((...((((((.......))))))...)))...)))..))). ( -37.70) >sp_ag.0 350400 120 + 2160267/1240-1360 GAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGCAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUU ....((.((((((((((......(((((.((....)).)))))...))))))))))))......(((......(((.(((...((((((.......))))))...)))...)))..))). ( -39.20) >consensus GAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUU ....(((((((..(((((((...(((((.((....)).)))))..))))))).......)))))))(((((............((((((.......)))))))))))............. (-37.42 = -36.58 + -0.84)

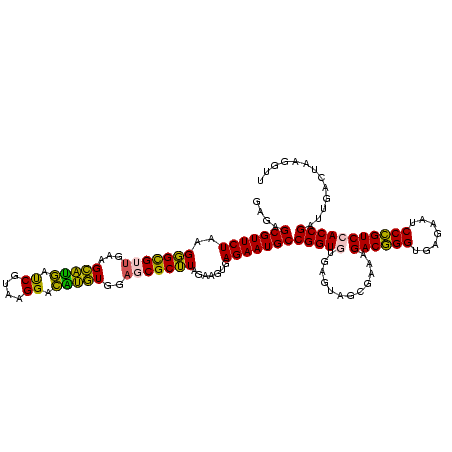
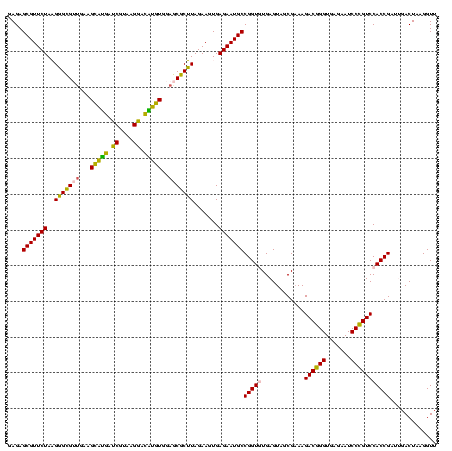
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.67 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -22.62 |
| Energy contribution | -20.46 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687306 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1240-1360 AACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUC ............((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))....... ( -25.90) >sy_au.0 694299 120 - 2809422/1240-1360 AACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUC ............((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))....... ( -25.90) >sy_sa.0 745546 120 + 2516575/1240-1360 AACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUC ............((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))....... ( -25.90) >sp_pn.0 16940 120 + 2038615/1240-1360 AACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCACUCCUCACGGUAUACCUUCAUCACACAUAGAACGCUCUC ........(((....(((((((((.......))))))...........((((((((((..........))))((....))........))))))........)))....)))........ ( -23.50) >sp_ag.0 350400 120 + 2160267/1240-1360 AACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUC ..............((((((((((.......))))))..(((((((.((((..((........))..)))).))...)))))...........))))((((........))))....... ( -25.60) >consensus AACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUC .............(((((((((((.......))))))....(.....).)))))..........(((((((.((....((((((.....)).)))).......)).)))))))....... (-22.62 = -20.46 + -2.16) # Strand winner: forward (1.00)

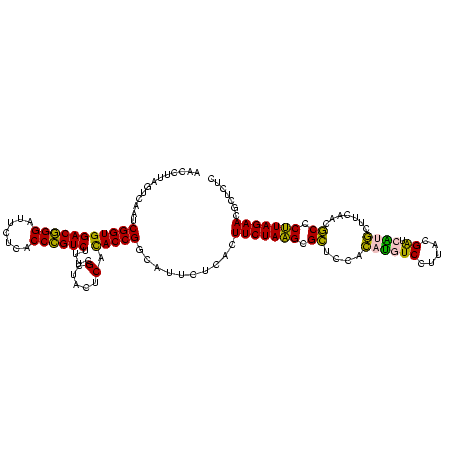

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -33.14 |
| Energy contribution | -32.34 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1280-1400 AGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACA .(((.((((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).))))))...............).).)))... ( -34.50) >sy_au.0 694299 120 - 2809422/1280-1400 AGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACA .(((.((((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).))))))...............).).)))... ( -34.50) >sy_sa.0 745546 120 + 2516575/1280-1400 AGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACA .(((.((((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).))))))...............).).)))... ( -34.50) >sp_pn.0 16940 120 + 2038615/1280-1400 AGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGC .(((...((((((.((((((.(((......))))))))).....))))))...(((.(((((((.......))))))).)))........))).((((..........))))........ ( -34.60) >sp_ag.0 350400 120 + 2160267/1280-1400 AGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGC .((..(.((((((.((((((.(((......))))))))).....))))))...(((((((((((.......)))))))............))))((((..........)))))..))... ( -34.50) >consensus AGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACA .(((.((((((((.((((((.(((......))))))))).....))))))...(((((((((((.......))))))....(.....).)))))................).).)))... (-33.14 = -32.34 + -0.80) # Strand winner: forward (1.00)

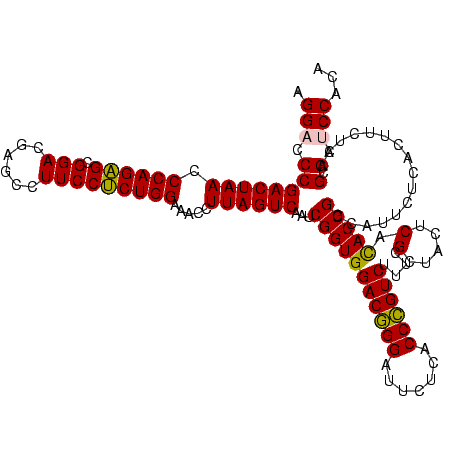

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -45.26 |
| Consensus MFE | -46.64 |
| Energy contribution | -44.40 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.03 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1320-1440 AAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUG ...((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...))))......... ( -46.10) >sy_au.0 694299 120 - 2809422/1320-1440 AAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUG ...((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...))))......... ( -46.10) >sy_sa.0 745546 120 + 2516575/1320-1440 AAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAUAGGCGUAGGCGAUGGAUAACAGGUUG ...((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...))))......... ( -46.10) >sp_pn.0 16940 120 + 2038615/1320-1440 AAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUG ...((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))......))).. ( -44.00) >sp_ag.0 350400 120 + 2160267/1320-1440 CAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUG ...((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))......))).. ( -44.00) >consensus AAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAAAGGCGUAGGCGAUGGAUAACAGGUUG ...((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))..((((...(((.(.(((....))).).)))...))))....))).. (-46.64 = -44.40 + -2.24)

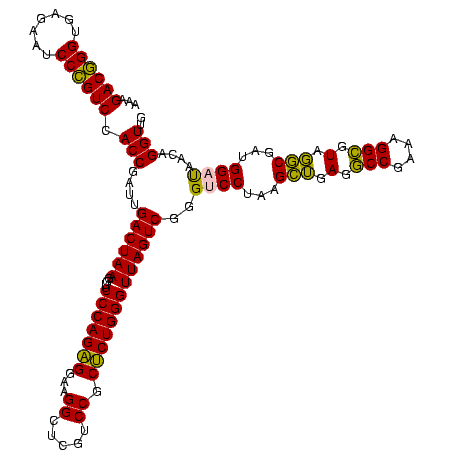
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -37.04 |
| Consensus MFE | -35.98 |
| Energy contribution | -34.46 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1320-1440 CAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUU ..(((.(((.(((...((....(((....)))....))...)))...((((((.((((((.(((......))))))))).....))))))))).)))(((((((.......))))))).. ( -36.70) >sy_au.0 694299 120 - 2809422/1320-1440 CAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUU ..(((.(((.(((...((....(((....)))....))...)))...((((((.((((((.(((......))))))))).....))))))))).)))(((((((.......))))))).. ( -36.70) >sy_sa.0 745546 120 + 2516575/1320-1440 CAACCUGUUAUCCAUCGCCUACGCCUAUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUU ..(((.(((.(((...((....(((....)))....))...)))...((((((.((((((.(((......))))))))).....))))))))).)))(((((((.......))))))).. ( -36.70) >sp_pn.0 16940 120 + 2038615/1320-1440 CAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUU ........(((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))))))((((........)))).. ( -37.40) >sp_ag.0 350400 120 + 2160267/1320-1440 CAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUG .........((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...))))))))(((((........))))). ( -37.70) >consensus CAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUU ..(((.....(((...((....(((....)))....))...)))...((((((.((((((.(((......))))))))).....))))))....)))(((((((.......))))))).. (-35.98 = -34.46 + -1.52) # Strand winner: forward (1.00)

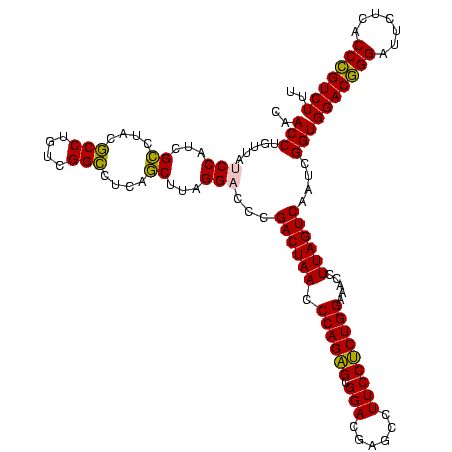

| Location | 973,169 – 973,287 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.82 |
| Mean single sequence MFE | -34.70 |
| Consensus MFE | -24.54 |
| Energy contribution | -22.54 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954854 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 118 + 2685015/1480-1600 GACGCAGUAGGAUAGGCGAAGCGUACGAUUGGAU-UGUACGUCUAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCC ..((((((..(((..((..((((((((((...))-)))))).))..)).........((((((..((....))..))))))-.....)))..))))))..((..(.((....)).)..)) ( -32.50) >sy_au.0 694299 119 - 2809422/1480-1600 GACGCAGUAGGAUAGGCGAAGCGUGCGAUUGGAU-UGCACGUCUAAGCAGUAAGGCUGAGUAUUAGGCAAAUCCGGUACUCGUUAAGGCUGAGCUGUGAUGGGGAGAAGACAUUGUGUCU ((((((((.((((..((...(((((((((...))-)))))))((((.(((.....)))....))))))..))))....(((((((.(((...))).)))))))........)))))))). ( -41.30) >sy_sa.0 745546 118 + 2516575/1480-1600 GACGCAGUAGGAUAGGCGAAGCGUACGAUUGGAU-UGUACGUCCAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCC ..(((..........)))..(((((((((...))-)))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..)))(((((.(....).))))) ( -33.70) >sp_pn.0 16940 119 + 2038615/1480-1600 GACGCAGUAGGCUAACUAAAGCAGACGAUUGGAAGAGUCUGUCUAAGCAGUGAGGUGUGAAUUGAGUCAAAUGCUUAAUUCUAUAACAUUGAGCUGUGAUGGGGAGC-GAAGUUUAGUAG ..........(((..(((..((((((..........))))))....(((((..((((((((((((((.....)))))))))....)))))..)))))..)))..)))-............ ( -31.80) >sp_ag.0 350400 119 + 2160267/1480-1600 GACGCAGUAGGCUAACUAAACCAGACGAUUGGAAGUGUCUGGUCAAACAGUGAGGUGUGAUAUGAGUCAAAUGCUUAUAUCUUUAACAUUGAGCUGUGACGAGGAGC-GAAGUUUAGUAG ...((.....))..(((((((((((((........))))))(((..(((((..((((((((((((((.....)))))))))....)))))..)))))))).......-...))))))).. ( -34.20) >consensus GACGCAGUAGGAUAGGCGAAGCGUACGAUUGGAU_UGUACGUCUAAGCAGUGAGGUUGAGUAUUAGGCAAAUCCGGAACUC_UUAAGAUUGAGCUGUGAUGGGGAGA_GAAAUUGUGUCC ((((((((............(((((((........)))))))(((.(((((..(((((((((((.((.....))))))))).....))))..)))))..))).........)))))))). (-24.54 = -22.54 + -2.00)



| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -31.28 |
| Energy contribution | -31.84 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1680-1800 AGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUG ..((..(((((((.(((..............))).)))))))...))(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).))). ( -39.44) >sy_au.0 694299 120 - 2809422/1680-1800 AGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUG ..((..(((((((.(((..............))).)))))))...))(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).))). ( -38.74) >sy_sa.0 745546 120 + 2516575/1680-1800 AGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGGAGAGCCGCAGUG ..((..(((((((.(((..............))).)))))))...))(((((((......(((((....(....).....)))))..((((..((((....))))..)))))))).))). ( -38.74) >sp_pn.0 16940 109 + 2038615/1680-1800 AGUCGAGGCGAGUAGCCUCAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCU-----GACUUUAAGUC------AGCCGCAGUG ....(((((.....))))).....(((((((....))))))).....(((((((......(((((....(....).....)))))..(-----(((.....)))------))))).))). ( -42.60) >sp_ag.0 350400 109 + 2160267/1680-1800 AGUCGAGGCGAGUAGCCUCAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCU-----GAUUUUAGAUC------AGCCGCAGUG .((((((((.....))))).(((.(((((((....))))))).....))).)))......(((((....(....).....)))))(((-----((((...))))------)))....... ( -42.90) >consensus AGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAAAGCCCAG_AGAGCCGCAGUG ........((((...(((......((((((......)))))).)))..))))((......(((((.....(((....))))))))(((((((.((((....)))).))))))).)).... (-31.28 = -31.84 + 0.56)

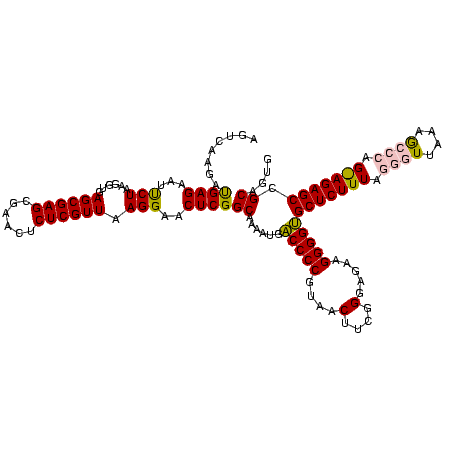
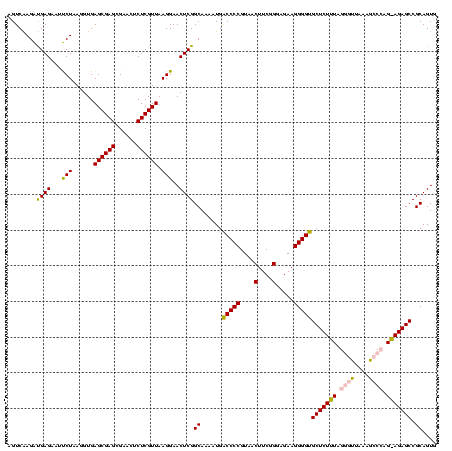
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -40.86 |
| Consensus MFE | -32.92 |
| Energy contribution | -35.16 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.907381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1760-1880 GGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCC ((((((((((((.((((....)))).)))))).)))......(((((...(((..((((.((((.......(((....)))....(((....))))))).))))..)))...)))))))) ( -45.30) >sy_au.0 694299 120 - 2809422/1760-1880 GGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCC ((((((((((((.((((....)))).)))))).)))......(((((...(((..((((.((((.......(((....)))....(((....))))))).))))..)))...)))))))) ( -44.60) >sy_sa.0 745546 120 + 2516575/1760-1880 GGGGUGCUCUUUAGGGUUAACGCCCAGGAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCC (((((((((((..((((....))))..))))).)))......(((((...(((..((((.((((.......(((....)))....(((....))))))).))))..)))...)))))))) ( -44.60) >sp_pn.0 16940 109 + 2038615/1760-1880 GGGGUGCU-----GACUUUAAGUC------AGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCC .(((((((-----(((.....)))------))).((.((((((((((....))..))))))))......(((..((((.(((...(((....)))...))).)))).)))..))..)))) ( -34.70) >sp_ag.0 350400 109 + 2160267/1760-1880 GGGGCGCU-----GAUUUUAGAUC------AGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCC .(((((((-----((((...))))------))).((.((((((((((....))..))))))))......(((..((((.(((...(((....)))...))).)))).)))..))..)))) ( -35.10) >consensus GGGGUGCUCUUUAGGGUUAAAGCCCAG_AGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCC ((((((((((((.((((....)))).)))))).)))......(((((.........(((((.....)))))..(((((((((...(((....)))...))..)))))))...)))))))) (-32.92 = -35.16 + 2.24)

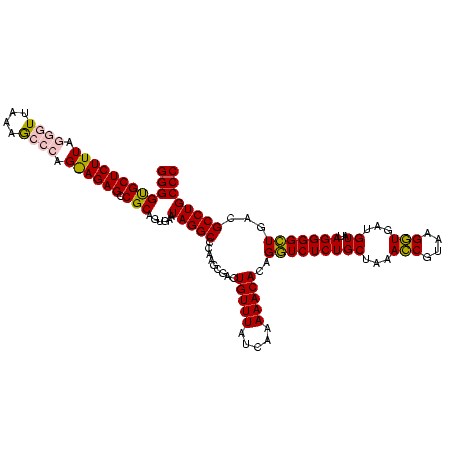
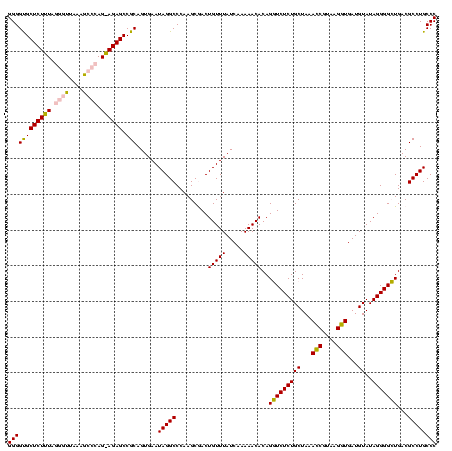
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -40.10 |
| Energy contribution | -41.46 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/1840-1960 GCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUA ((...(((....)))...))((((..((....(((.(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))))).))..)))). ( -43.20) >sy_au.0 694299 120 - 2809422/1840-1960 GCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUA ((...(((....)))...))((((..((....(((.(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))))).))..)))). ( -43.20) >sy_sa.0 745546 120 + 2516575/1840-1960 GCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUA ((...(((....)))...))((((..((....(((.(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))))).))..)))). ( -43.20) >sp_pn.0 16940 120 + 2038615/1840-1960 GCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUA ((...(((....)))...))......(((.(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).))).)))... ( -40.70) >sp_ag.0 350400 120 + 2160267/1840-1960 GCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUA ((...(((....)))...))......(((.(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).))).)))... ( -40.70) >consensus GCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUA ((...(((....)))...))((((......(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).)))..)))). (-40.10 = -41.46 + 1.36)


| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -42.60 |
| Energy contribution | -37.96 |
| Covariance contribution | -4.64 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.25 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990537 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2120-2240 UGUAGCCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUA ..............((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..)))).. ( -45.50) >sy_au.0 694299 120 - 2809422/2120-2240 UGUAGCCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUA ..............((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..)))).. ( -45.50) >sy_sa.0 745546 120 + 2516575/2120-2240 UGUAGUCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUA ..............(((((((((((((((((.(((.......))).))((((....(((((((((....))))))))).))))(..(((((....)))))..)))))))))))))))).. ( -49.70) >sp_pn.0 16940 120 + 2038615/2120-2240 UGCAGUUUGAUAUUGAGUGUCUGUACCACAUGUACAGGAUAGGUAGGAGUCUAAGAGAUCGGGACGCCAGUUUCGAAGGAGACGCUGUUGGGAUACUACCCUUGUGUUAUGGCCACUCUA ..............((((((((((((.....)))))))...(((((...(((((.((.(((..((....))..))).(....).)).)))))...))))).............))))).. ( -36.90) >sp_ag.0 350400 120 + 2160267/2120-2240 UGCAGUUUGAUAUUGAGUAUCUGUACCACAUGUACAGGAUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUA ..............((((((((((((.....)))))))...(((((...(((..((((((((....)).(((((....))))))))))))))...))))).............))))).. ( -32.40) >consensus UGUAGUCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUAUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUA ..............((((((((((((((((.............(((..((((.....((((((((....))))))))..)))).)))..(((......))).)))))))))))))))).. (-42.60 = -37.96 + -4.64)


| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.83 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -29.94 |
| Energy contribution | -26.26 |
| Covariance contribution | -3.68 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2120-2240 UAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGGCUACA .....((((..(((((((.(((......)))......((((..((((.(((....))).)))).....))))..................)))))))..(.....).......))))... ( -32.10) >sy_au.0 694299 120 - 2809422/2120-2240 UAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGGCUACA .....((((..(((((((.(((......)))......((((..((((.(((....))).)))).....))))..................)))))))..(.....).......))))... ( -32.10) >sy_sa.0 745546 120 + 2516575/2120-2240 UAGAAAUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUCUCAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGACUACA ..(((((.(..((((((..(((......)))......((((..((((.(((....))).)))).....))))...................))))))..).))))).............. ( -30.00) >sp_pn.0 16940 120 + 2038615/2120-2240 UAGAGUGGCCAUAACACAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGGCGUCCCGAUCUCUUAGACUCCUACCUAUCCUGUACAUGUGGUACAGACACUCAAUAUCAAACUGCA ..(((((............((((((........(((..((......))..)))..(((...........)))..))))))...((((((.....)))))).))))).............. ( -27.10) >sp_ag.0 350400 120 + 2160267/2120-2240 UAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGUACAUGUGGUACAGAUACUCAAUAUCAAACUGCA ..(((((((((((((....(((......)))....(((((............)))))...)))))....)))...........((((((.....)))))).))))).............. ( -31.00) >consensus UAGAAAGCCAACACAGCAAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUAUCAAAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGACUACA ..(((((.(..(((((((.(((......)))......((((..((((.(((....))).)))).....))))..................)))))))..).))))).............. (-29.94 = -26.26 + -3.68) # Strand winner: forward (1.00)

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -43.54 |
| Consensus MFE | -39.24 |
| Energy contribution | -34.24 |
| Covariance contribution | -5.00 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908423 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2160-2280 AGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGG ........(((((....((((((((....)))))))).)))))((((((((....)))))...((((((((((....)))))((.((((((.....))))))))..)))))....))).. ( -48.40) >sy_au.0 694299 120 - 2809422/2160-2280 AGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGG ........(((((....((((((((....)))))))).)))))((((((((....)))))...((((((((((....)))))((.((((((.....))))))))..)))))....))).. ( -48.40) >sy_sa.0 745546 120 + 2516575/2160-2280 AGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGG ........((((....(((((((((....))))))))).))))...(((((....)))))..(((((((((((....)))))((.((((((.....))))))))..))))))........ ( -45.50) >sp_pn.0 16940 120 + 2038615/2160-2280 AGGUAGGAGUCUAAGAGAUCGGGACGCCAGUUUCGAAGGAGACGCUGUUGGGAUACUACCCUUGUGUUAUGGCCACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGACGG .(((((...(((((.((.(((..((....))..))).(....).)).)))))...)))))....(((((.(((.(((........((((((.....))))))(....)))))))))))). ( -39.40) >sp_ag.0 350400 120 + 2160267/2160-2280 AGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGACGG .(((((...(((..((((((((....)).(((((....))))))))))))))...)))))....(((((.(((.(((........((((((.....))))))(....)))))))))))). ( -36.00) >consensus AGGUAGGAGCCUAUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCCUGGUGGGAGACAGUGUCAGACGG .(((.(((((((.....((((((((....))))))))..))))((.(((((....)))))...))...........))).)))..((((((.....))))))(....)............ (-39.24 = -34.24 + -5.00)

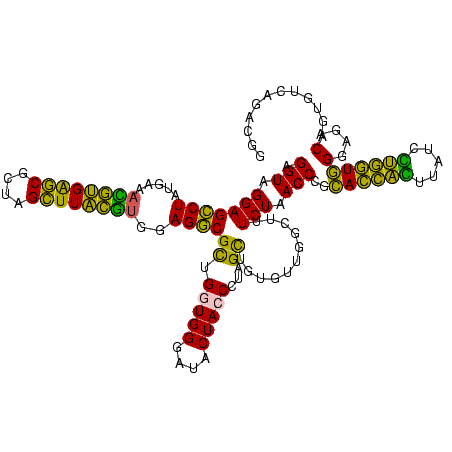
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.33 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -32.34 |
| Energy contribution | -28.06 |
| Covariance contribution | -4.28 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2160-2280 CCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCU ..((.((...(((.(((.((((((.....)))))).))).)))..(((.......))).(((......)))..))))((((..((((.(((....))).)))).....))))........ ( -34.70) >sy_au.0 694299 120 - 2809422/2160-2280 CCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCU ..((.((...(((.(((.((((((.....)))))).))).)))..(((.......))).(((......)))..))))((((..((((.(((....))).)))).....))))........ ( -34.70) >sy_sa.0 745546 120 + 2516575/2160-2280 CCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUCUCAGGCUCCUACCU ...((((((.(((.....((((((.....)))))))))))))))..........((.(((((......)))......((((..((((.(((....))).)))).....)))))))).... ( -31.70) >sp_pn.0 16940 120 + 2038615/2160-2280 CCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACACAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGGCGUCCCGAUCUCUUAGACUCCUACCU ..(((((((.(((((((.((((((.....)))))).))).....(((.......)))..(((......))).)))).))))...(((..((....))..))).......)))........ ( -31.60) >sp_ag.0 350400 120 + 2160267/2160-2280 CCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCU ..........(((.(((.((((((.....)))))).))).)))((.(((((((((....(((......)))....(((((............)))))...)))))....)))).)).... ( -33.10) >consensus CCGUCUGACACUGUCUCCCACCACGAUAACUGGUGCGGGUUAGAAAGCCAACACAGCAAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUAUCAAAGGCUCCUACCU ..........(((.(((.((((((.....)))))).))).)))................(((......)))......((((..((((.(((....))).)))).....))))........ (-32.34 = -28.06 + -4.28) # Strand winner: forward (1.00)


| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -43.76 |
| Consensus MFE | -46.70 |
| Energy contribution | -43.34 |
| Covariance contribution | -3.36 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.41 |
| Structure conservation index | 1.07 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2240-2360 ACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCA .(((.((((((.....))))))(....)...(((((((.....))))))).)))..(..(((((((...........)))))))..).............(((((((.....))))))). ( -44.10) >sy_au.0 694299 120 - 2809422/2240-2360 ACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCA .(((.((((((.....))))))(....)...(((((((.....))))))).)))..(..(((((((...........)))))))..).............(((((((.....))))))). ( -44.10) >sy_sa.0 745546 120 + 2516575/2240-2360 ACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCA .(((.((((((.....))))))(....)...(((((((.....))))))).)))..(..(((((((...........)))))))..)...(((((((...........)))))))..... ( -41.70) >sp_pn.0 16940 120 + 2038615/2240-2360 ACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCG .((((((((((.....))))))(....)...(((.(((.....))).)))))))..((.(((((((...........))))))).)).............(((((((.....))))))). ( -42.60) >sp_ag.0 350400 120 + 2160267/2240-2360 ACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCG ..((.((((((.....)))))))).(((.(..((..((...((((((((..(((((((.(((((((...........))))))).))....))))).))))))))))..))...).))). ( -46.30) >consensus ACCCGCACCACUUAUCCUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCA .((((((((((.....))))))(....)...(((((((.....)))))))))))..((.(((((((...........))))))).)).............(((((((.....))))))). (-46.70 = -43.34 + -3.36)



| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -38.94 |
| Consensus MFE | -37.04 |
| Energy contribution | -34.36 |
| Covariance contribution | -2.68 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2240-2360 UGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGU .(((((.........)))))((.(((......((..(((((((...........)))))))..)))))))........((((((..(((...)))....(((((.....))))))))))) ( -38.40) >sy_au.0 694299 120 - 2809422/2240-2360 UGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGU .(((((.........)))))((.(((......((..(((((((...........)))))))..)))))))........((((((..(((...)))....(((((.....))))))))))) ( -38.40) >sy_sa.0 745546 120 + 2516575/2240-2360 UGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGU .(((((.........)))))...(....)....(..(((((((...........)))))))..)((((..((((...........)))).........((((((.....)))))).)))) ( -33.20) >sp_pn.0 16940 120 + 2038615/2240-2360 CGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGU .........((((.....((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))..........((((((.....)))))))))). ( -40.80) >sp_ag.0 350400 120 + 2160267/2240-2360 CGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGU .(((((.....))....(((((((((.......((((((((((...........))))))....))))(((.....)))))).))))))...)))(((((((((.....)))))).))). ( -43.90) >consensus UGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCUGACACUGUCUCCCACCACGAUAACUGGUGCGGGU .(((((.........)))))((.(((..........(((((((...........)))))))....)))))........((((....(((...)))...((((((.....)))))).)))) (-37.04 = -34.36 + -2.68) # Strand winner: forward (1.00)

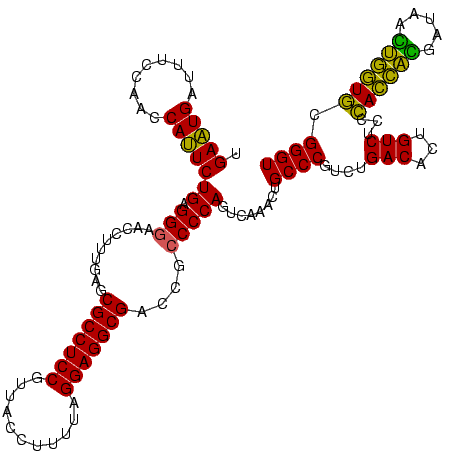
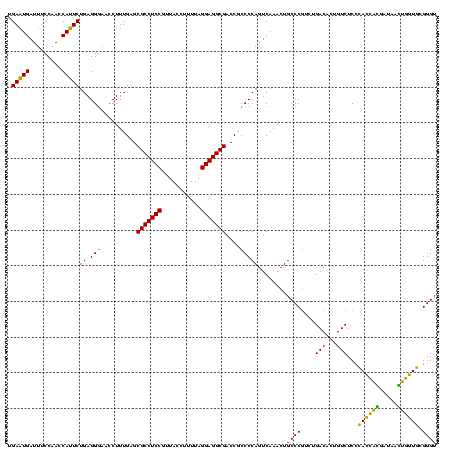
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -43.56 |
| Consensus MFE | -43.04 |
| Energy contribution | -41.92 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.18 |
| SVM RNA-class probability | 0.999977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2280-2400 GCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUA .(((.....)))....((((((((((...........))))))((....(((((((((..(((((((.....))))))).....((((....)))).)))))))))....))..)))).. ( -43.90) >sy_au.0 694299 120 - 2809422/2280-2400 GCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUA .(((.....)))....((((((((((...........))))))((....(((((((((..(((((((.....))))))).....((((....)))).)))))))))....))..)))).. ( -43.90) >sy_sa.0 745546 120 + 2516575/2280-2400 GCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUA .(((.....)))..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))........ ( -41.40) >sp_pn.0 16940 120 + 2038615/2280-2400 GCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUA ((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....)))))))((.....)).........)))))))))))))))........ ( -43.30) >sp_ag.0 350400 120 + 2160267/2280-2400 GCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUA ((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....)))))))......((((....)))))))))))))))))))........ ( -45.30) >consensus GCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUA ((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....(((......))).)))))))))))))))........ (-43.04 = -41.92 + -1.12)

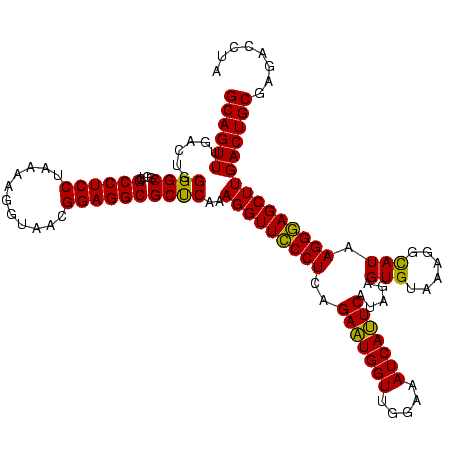
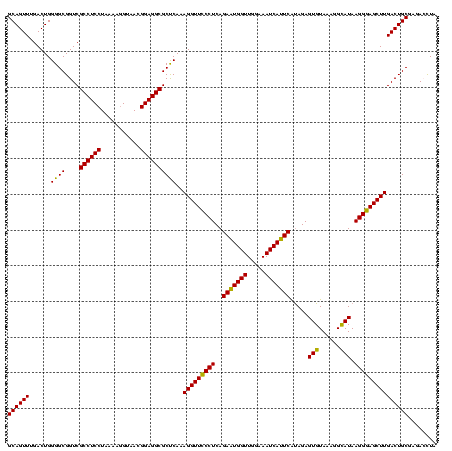
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.33 |
| Mean single sequence MFE | -33.01 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.38 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997772 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2280-2400 UAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGC ..((((..((..(((((.(((((((((......))......(((((.........))))))))))))...)))))))((((((...........))))))))))....(((.....))). ( -33.90) >sy_au.0 694299 120 - 2809422/2280-2400 UAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGC ..((((..((..(((((.(((((((((......))......(((((.........))))))))))))...)))))))((((((...........))))))))))....(((.....))). ( -33.90) >sy_sa.0 745546 120 + 2516575/2280-2400 UAAGUCUCGCAGUCAAGCUUCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGC ........((.(((..((((.......((((..........(((((.........))))).))))........))))((((((...........))))))))).))..(((.....))). ( -28.46) >sp_pn.0 16940 120 + 2038615/2280-2400 UAGCUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGC ........(((((...(((((((((........(.....).(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...))))) ( -33.30) >sp_ag.0 350400 120 + 2160267/2280-2400 UAGCUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGC ........(((((...(((((((((................(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...))))) ( -35.49) >consensus UAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGC ........((.(((..((((.......((((((........(((((.........))))))))))).......))))((((((...........))))))))).))..(((.....))). (-30.90 = -30.38 + -0.52) # Strand winner: forward (1.00)

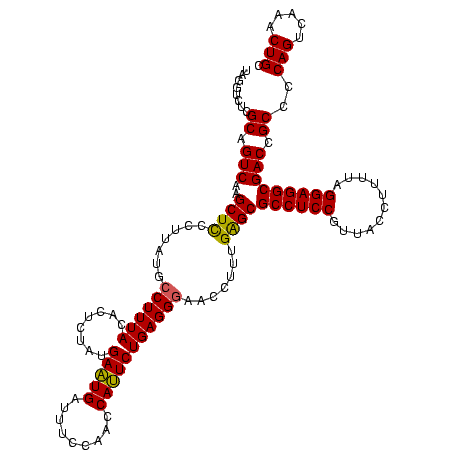

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.00 |
| Mean single sequence MFE | -40.84 |
| Consensus MFE | -39.10 |
| Energy contribution | -36.78 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2320-2440 GGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGU ((((((...(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))......))))..))... ( -39.40) >sy_au.0 694299 120 - 2809422/2320-2440 GGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGU ((((((...(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))......))))..))... ( -39.40) >sy_sa.0 745546 120 + 2516575/2320-2440 GGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGU ((((((...(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..((((....))))..)))).(((....)))......))))..))... ( -38.20) >sp_pn.0 16940 120 + 2038615/2320-2440 GGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGU ...((((..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....)))))))............. ( -42.60) >sp_ag.0 350400 120 + 2160267/2320-2440 GGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGU ...((((..(((((((((..(((((((.....)))))))......((((....)))))))))))))..((((..(((......)))..)))).(((....)))))))............. ( -44.60) >consensus GGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGU ...(((...(((((((((..(((((((.....))))))).....(((......))).)))))))))..((((..(((......)))..)))).(((....)))((((.....).)))))) (-39.10 = -36.78 + -2.32)


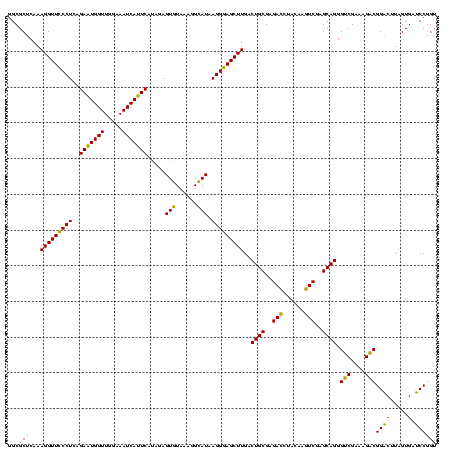
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -36.84 |
| Energy contribution | -35.72 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.995080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2360-2480 UAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCU ..(((((....(((.((((...((((((((............))))))))...(((....)))...))))....(((((((....))).))))...)))..))))).............. ( -38.50) >sy_au.0 694299 120 - 2809422/2360-2480 UAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCU ..(((((....(((.((((...((((((((............))))))))...(((....)))...))))....(((((((....))).))))...)))..))))).............. ( -38.50) >sy_sa.0 745546 120 + 2516575/2360-2480 UAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCU ..(((((....(((.((((...((((((((............))))))))...(((....)))...))))....(((((((....))).))))...)))..))))).............. ( -38.50) >sp_pn.0 16940 120 + 2038615/2360-2480 CAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCU ..((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).............. ( -40.10) >sp_ag.0 350400 120 + 2160267/2360-2480 UAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCU ..((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).............. ( -40.10) >consensus UAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCU ..((((............(((.(((...((((..(((......)))..)))).(((....)))......)))..)))((((((.((........)))))))))))).............. (-36.84 = -35.72 + -1.12)

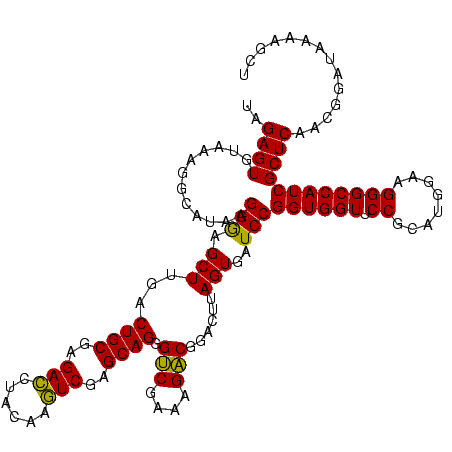
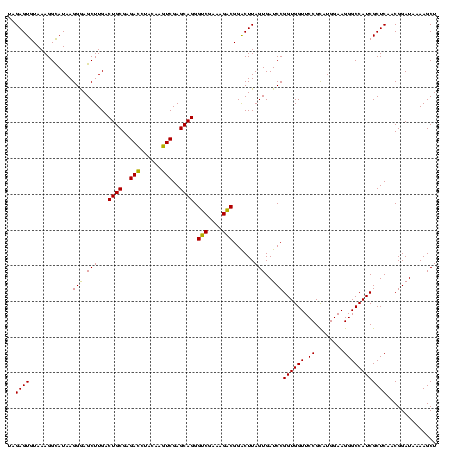
| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -56.20 |
| Consensus MFE | -56.52 |
| Energy contribution | -56.04 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.37 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2520-2640 UCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGA .(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....)))))... ( -55.40) >sy_au.0 694299 120 - 2809422/2520-2640 UCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGA .(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....)))))... ( -55.40) >sy_sa.0 745546 120 + 2516575/2520-2640 UCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGA .(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....)))))... ( -55.40) >sp_pn.0 16940 120 + 2038615/2520-2640 UCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGA .(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....)))))))))..))....)))))... ( -57.40) >sp_ag.0 350400 120 + 2160267/2520-2640 UCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGA .(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....)))))))))..))....)))))... ( -57.40) >consensus UCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGA .(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....)))))... (-56.52 = -56.04 + -0.48)

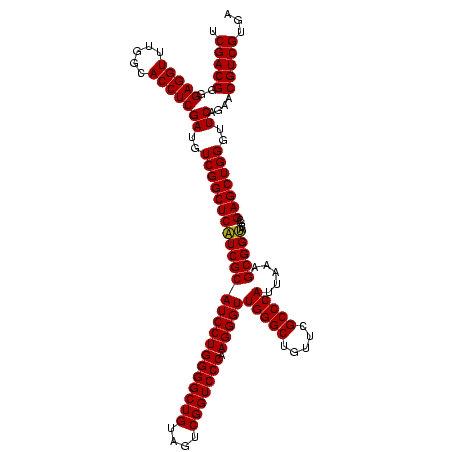

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -42.30 |
| Consensus MFE | -42.62 |
| Energy contribution | -42.14 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.56 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2520-2640 UCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGA ...(((((...........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).......(((((.....)))))..))))). ( -41.90) >sy_au.0 694299 120 - 2809422/2520-2640 UCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGA ...(((((...........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).......(((((.....)))))..))))). ( -41.90) >sy_sa.0 745546 120 + 2516575/2520-2640 UCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGA ...(((((...........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).......(((((.....)))))..))))). ( -41.90) >sp_pn.0 16940 120 + 2038615/2520-2640 UCACGACGUUCUGAACCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGA ...(((((...........((((((((..........(((((.....)))))...((((((.(........).))))))..))).))))).......(((((.....)))))..))))). ( -42.90) >sp_ag.0 350400 120 + 2160267/2520-2640 UCACGACGUUCUGAACCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGA ...(((((...........((((((((..........(((((.....)))))...((((((.(........).))))))..))).))))).......(((((.....)))))..))))). ( -42.90) >consensus UCACGACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGA ...(((((...........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).))))).......(((((.....)))))..))))). (-42.62 = -42.14 + -0.48) # Strand winner: forward (1.00)

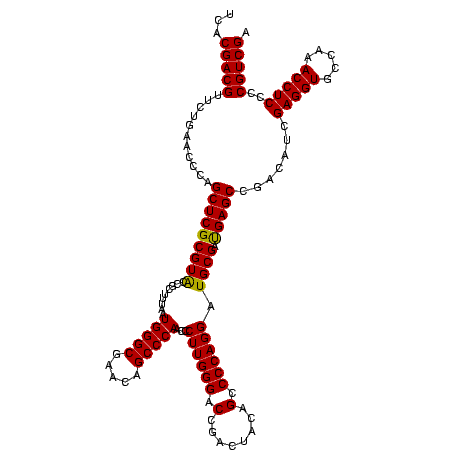

| Location | 973,169 – 973,289 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -49.60 |
| Consensus MFE | -46.30 |
| Energy contribution | -48.82 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 120 + 2685015/2560-2680 UCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAAAUUUG (((((((((((....))))))).)))).....((...(((((((....((((..(....)..(((..(.(((((((....))).)))))..)))...)))).)))))))..))....... ( -48.20) >sy_au.0 694299 120 - 2809422/2560-2680 UCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAAAUUUG (((((((((((....))))))).)))).....((...(((((((....((((..(....)..(((..(.(((((((....))).)))))..)))...)))).)))))))..))....... ( -48.20) >sy_sa.0 745546 120 + 2516575/2560-2680 UCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAAAUUUG (((((((((((....))))))).)))).....((...(((((((....((((..(....)..(((..(.(((((((....))).)))))..)))...)))).)))))))..))....... ( -48.20) >sp_pn.0 16940 120 + 2038615/2560-2680 UCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAAAUUUG (((((((((((....))))))).)))).(((((.....))))).....(((.(.((((((.((((..(.(((((((....))).)))).)..))))..).)))))).))).......... ( -51.70) >sp_ag.0 350400 120 + 2160267/2560-2680 UCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAAAUUUG (((((((((((....))))))).)))).(((((.....))))).....(((.(.((((((.((((..(.(((((((....))).)))).)..))))..).)))))).))).......... ( -51.70) >consensus UCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAAAUUUG (((((((((((....))))))).)))).(((((.....))))).....(((.(.((((((.((((..(.(((((((....))).)))).)..))))..).)))))).))).......... (-46.30 = -48.82 + 2.52)

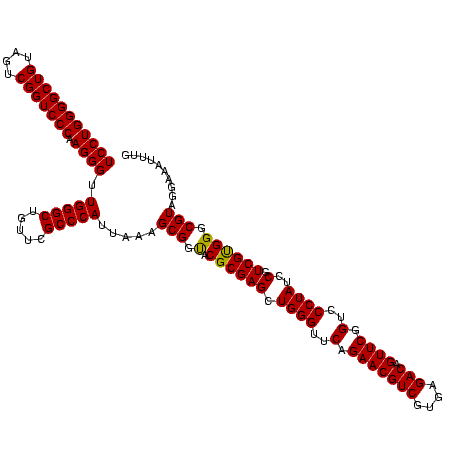

| Location | 973,169 – 973,285 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -24.52 |
| Energy contribution | -23.80 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 973169 116 + 2685015/2824-2944 AUUUCCCA--ACUUCGGUU--AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCGUGGUGACACGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAUU ........--.((((((((--........(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))............ ( -33.40) >sy_au.0 694299 116 - 2809422/2824-2944 AUUUCCCA--ACUUCGGUU--AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAA ........--.((((((((--........(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))............ ( -32.00) >sy_sa.0 745546 116 + 2516575/2824-2944 AUUUCCCA--ACUUCGGUU--AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUAGCGAUAUGUGUAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAU ........--.((((((((--........(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))............ ( -26.90) >sp_pn.0 16940 120 + 2038615/2824-2944 AUUUCCCAUGAUUAUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGAAGUGGAAGUGUGGCGACACAUGUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAG .....((.(((((((...((((........))))...)))))))))..((((((((....((....(((((....)))))...))(..(((.......)))..)...))))))))..... ( -30.20) >sp_ag.0 350400 120 + 2160267/2824-2944 AUUUCCCAUGAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGGAGUGUAAUUGUGGUGACACAUUUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAA ...((((.((((.....))))....(((((((((.......)))))....((((((((..((.(((.((((....)))).)))))...))))).)))...))))).)))........... ( -30.20) >consensus AUUUCCCA__ACUUCGGUU__AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCGUGGUGACACGUGUAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAA .........................(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))................ (-24.52 = -23.80 + -0.72)

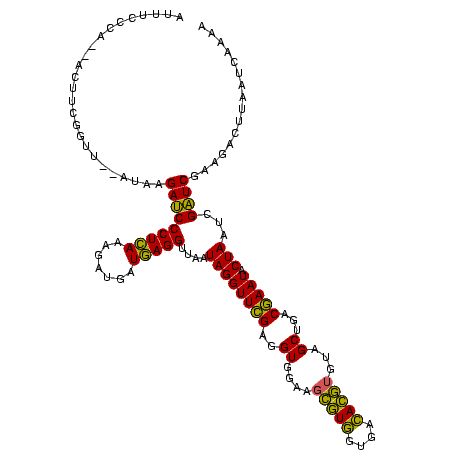
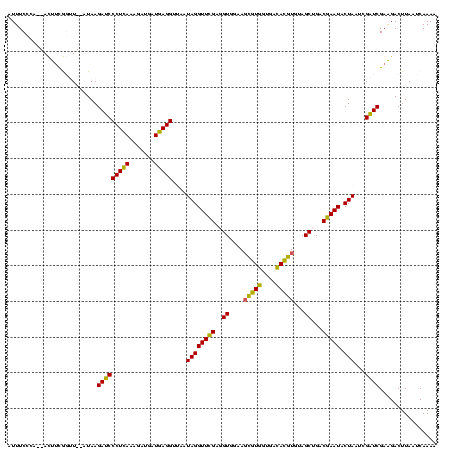
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 19:00:06 2006