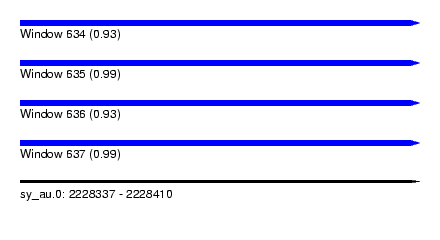
| Sequence ID | sy_au.0 |
|---|---|
| Location | 2,228,337 – 2,228,410 |
| Length | 73 |
| Max. P | 0.994826 |

| Location | 2,228,337 – 2,228,410 |
|---|---|
| Length | 73 |
| Sequences | 4 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -27.73 |
| Energy contribution | -26.10 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
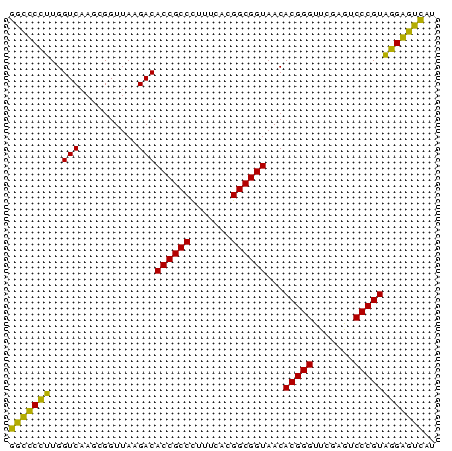
>sy_au.0 2228337 73 + 2809422 AUGACUCCUACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCC ....((((((((((.......)))))..(((((((.......))))))).................))))).. ( -25.10) >sy_ha.0 1800022 73 - 2685015 GUGACUCCUACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCC ....((((((((((.......)))))..(((((((.......))))))).................))))).. ( -25.10) >sy_sa.0 1767743 73 - 2516575 AUGACCCCGACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCACGGGGCC ....(((((.((((((.((....)).))).))).)(((...(((((((......)))))))...))))))).. ( -29.90) >sp_pn.0 1697429 73 + 2038615 AUAGUCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAAGGCGGUGUCUUAACCCCUUGACCAACGGACC ...(((((((((((.......)))))..(((((((.......))))))).................)))))). ( -28.40) >consensus AUGACCCCUACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCC ....((((((((((.......)))))..(((((((.......))))))).................))))).. (-27.73 = -26.10 + -1.63)

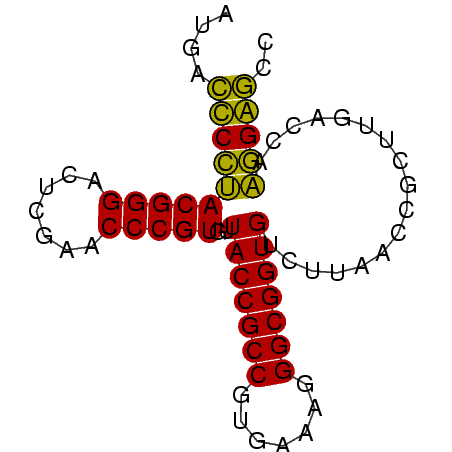

| Location | 2,228,337 – 2,228,410 |
|---|---|
| Length | 73 |
| Sequences | 4 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -31.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.09 |
| Structure conservation index | 1.06 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228337 73 + 2809422 GGCUCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAU (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. ( -29.10) >sy_ha.0 1800022 73 - 2685015 GGCUCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAC (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. ( -29.10) >sy_sa.0 1767743 73 - 2516575 GGCCCCGUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUCGGGGUCAU ((((((((((....(((((......)))))....))))(((((..((.((....)).)).))))))))))).. ( -32.70) >sp_pn.0 1697429 73 + 2038615 GGUCCGUUGGUCAAGGGGUUAAGACACCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAU (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. ( -28.80) >consensus GGCCCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAU (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. (-31.70 = -29.70 + -2.00) # Strand winner: reverse (0.91)

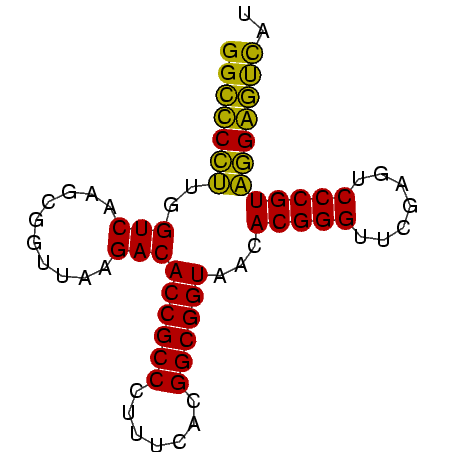
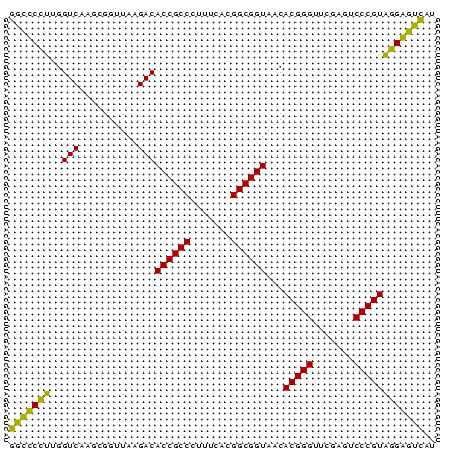
| Location | 2,228,337 – 2,228,410 |
|---|---|
| Length | 73 |
| Sequences | 4 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -27.73 |
| Energy contribution | -26.10 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.931799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228337 73 + 2809422/0-73 AUGACUCCUACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCC ....((((((((((.......)))))..(((((((.......))))))).................))))).. ( -25.10) >sy_ha.0 1800022 73 - 2685015/0-73 GUGACUCCUACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCC ....((((((((((.......)))))..(((((((.......))))))).................))))).. ( -25.10) >sy_sa.0 1767743 73 - 2516575/0-73 AUGACCCCGACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCACGGGGCC ....(((((.((((((.((....)).))).))).)(((...(((((((......)))))))...))))))).. ( -29.90) >sp_pn.0 1697429 73 + 2038615/0-73 AUAGUCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAAGGCGGUGUCUUAACCCCUUGACCAACGGACC ...(((((((((((.......)))))..(((((((.......))))))).................)))))). ( -28.40) >consensus AUGACCCCUACGGGACUCGAACCCGUGUUACCGCCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCC ....((((((((((.......)))))..(((((((.......))))))).................))))).. (-27.73 = -26.10 + -1.63)

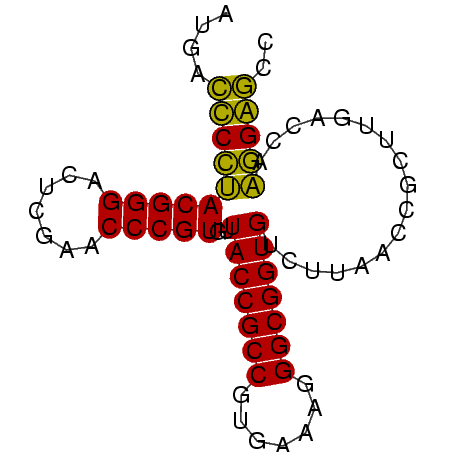
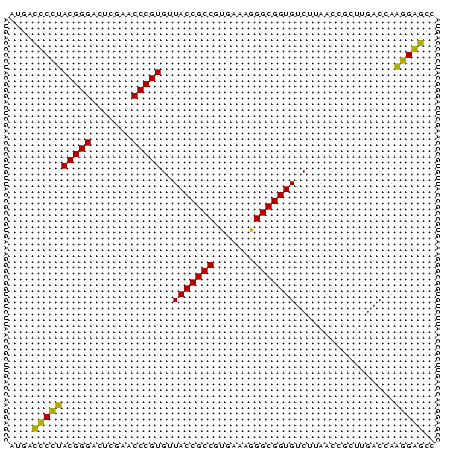
| Location | 2,228,337 – 2,228,410 |
|---|---|
| Length | 73 |
| Sequences | 4 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 89.95 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -31.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.09 |
| Structure conservation index | 1.06 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228337 73 + 2809422/0-73 GGCUCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAU (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. ( -29.10) >sy_ha.0 1800022 73 - 2685015/0-73 GGCUCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAC (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. ( -29.10) >sy_sa.0 1767743 73 - 2516575/0-73 GGCCCCGUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUCGGGGUCAU ((((((((((....(((((......)))))....))))(((((..((.((....)).)).))))))))))).. ( -32.70) >sp_pn.0 1697429 73 + 2038615/0-73 GGUCCGUUGGUCAAGGGGUUAAGACACCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAU (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. ( -28.80) >consensus GGCCCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAU (((((((..(((..........)))((((((.......))))))...(((((.......)))))))))))).. (-31.70 = -29.70 + -2.00) # Strand winner: reverse (0.91)
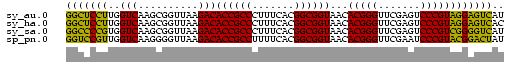
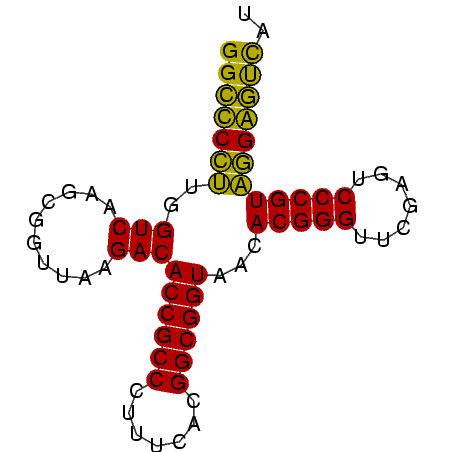
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:59:23 2006