
| Sequence ID | sy_au.0 |
|---|---|
| Location | 2,231,644 – 2,231,763 |
| Length | 119 |
| Max. P | 0.992059 |

| Location | 2,231,644 – 2,231,763 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.44 |
| Mean single sequence MFE | -42.82 |
| Consensus MFE | -32.90 |
| Energy contribution | -27.66 |
| Covariance contribution | -5.24 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
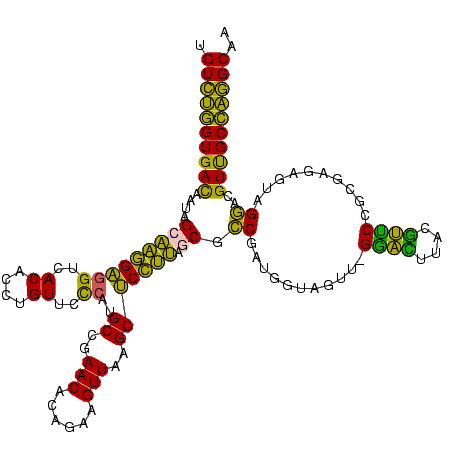
>sy_au.0 2231644 119 - 2809422/0-120 UGUCUGGUGACUAUAGCAAGGAGGUCACACCUGUUCCCAUGCCGAACACAGAAGUUAAGCUCCUUAGCGUCGAUGGUAGUC-GAACUUACGUUCCGCUAGAGUAGAACGUUGCCAGGCAG (((((((..((....(((((((((..((....))..))..((..(((......)))..))))))).)).(((((....)))-))......((((..(....)..))))))..))))))). ( -40.00) >sy_sa.0 2306446 119 + 2516575/0-120 UGUCUGGUGACAAUGGCAAGGAGGUCACACCUGUUCCCAUGCCGAACACAGAAGUUAAGCUCCUUAGCGCCGAUGGUAGUC-GGACUUACGUUCCGCAAGAGUAGGACGUUGCCAGGCAA (((((((..((..((((......))))..(((((((...(((.((((......((((((...)))))).(((((....)))-))......)))).))).)))))))..))..))))))). ( -43.70) >sy_ha.0 2495317 119 + 2685015/0-120 UGUCUGGUGACAAUGGCAAGGAGGUCACACCUGUUCUCAUGCCGAACACAGAAGUUAAGCUCCUUAGCGCCGAUGGUAGUU-GGAUUUACGUUCCGCUAGAGUAGGACGUUGCCAGGCAA (((((((..((..((((......))))..((((((((...((.((((......((((((...)))))).(((((....)))-))......)))).)).))))))))..))..))))))). ( -42.30) >sp_mu.0 2008906 120 - 2030921/0-120 UGUUAAGUGAUGAUAGCCUAGGGGAGACACCUGUAUCCAUGCCGAACACAGCAGUUAAGCCCUAGCACGCCUGAAGUAGUUGGGGGUUGCCCCCUGUGAGAUAGGGUCGUCGCUUAGCAA ((((((((((((((..(((((((((.((....)).)))...))...(((((..(.(((.(((((((((.......)).))))))).))).)..)))))...)))))))))))))))))). ( -47.00) >sp_ag.0 2064131 120 - 2160267/0-120 AGUUAAGUGAUAAUUGCCUAGGAGAUACACCUGUUCUCAUGCCGAACACAGAAGUUAAGCCCUAGAACGCCGGAAGUAGUUGGGGGUUGCCCCCUGUGAGAUAAGGUAGUCGCUUAGCAA .((((((((((...(((((..((((.((....))))))........(((((......((((((..(((..(....)..))).)))))).....))))).....))))))))))))))).. ( -41.10) >consensus UGUCUGGUGACAAUAGCAAGGAGGUCACACCUGUUCCCAUGCCGAACACAGAAGUUAAGCUCCUUAGCGCCGAUGGUAGUU_GGACUUACGUUCCGCGAGAGUAGGACGUUGCCAGGCAA .((((((((((....(((((((((..((....))..))..((..(((......)))..))))))).)).((...........((((....))))..........))..)))))))))).. (-32.90 = -27.66 + -5.24) # Strand winner: reverse (1.00)

| Location | 2,231,644 – 2,231,761 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.12 |
| Mean single sequence MFE | -29.86 |
| Consensus MFE | -18.20 |
| Energy contribution | -15.68 |
| Covariance contribution | -2.52 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
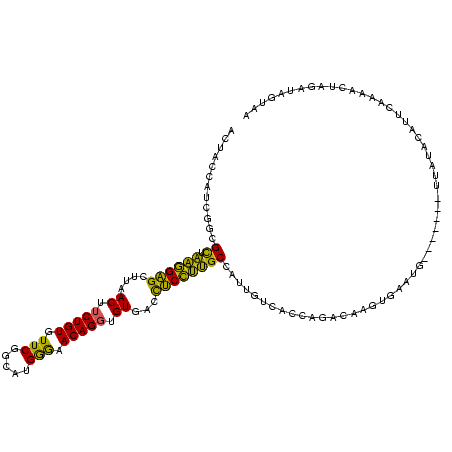
>sy_au.0 2231644 117 - 2809422/40-160 ACUACCAUCGACGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCUAUAGUCACCAGACAUAUGAAUGUA---AUUUAUACAUUCAAAACUAGAUAGUAA ..(((.(((...((.((((((.(((...(((((.(((......))))))))..))).))))))))....(((....)))...((((((((---.....))))))))......))).))). ( -30.30) >sy_sa.0 2306446 120 + 2516575/40-160 ACUACCAUCGGCGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCCAUUGUCACCAGACAAUGAAAUGUAUGAAAUUAUACAUUCAAAACUAGAUAGUAA ..(((.(((...((.((((((.(((...(((((.(((......))))))))..))).))))))))(((((((....))))))).((((((((....))))))))........))).))). ( -32.50) >sy_ha.0 2495317 114 + 2685015/40-160 ACUACCAUCGGCGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGAGAACAGGUGUGACCUCCUUGCCAUUGUCACCAGACAAGAGAAUG------AUUAUACAUUCAAAACUAGAUAGUAA ..(((.(((((((...(((((.(((...(((((.(((.....))).)))))..))).))))))))).(((((....)))))..(((((------......))))).......))).))). ( -31.20) >sp_mu.0 2008906 112 - 2030921/40-160 ACUACUUCAGGCGUGCUAGGGCUUAACUGCUGUGUUCGGCAUGGAUACAGGUGUCUCCCCUAGGCUAUCAUCACUUAACAAGCGUGA--------UCGCUCACUCACGAUUGAAUAGCUA .(((.(((((.((((((((((....((..((((((((.....))))))))..))...))))))((.((((.(.((.....)).))))--------).)).....)))).))))))))... ( -32.10) >sp_ag.0 2064131 113 - 2160267/40-160 ACUACUUCCGGCGUUCUAGGGCUUAACUUCUGUGUUCGGCAUGAGAACAGGUGUAUCUCCUAGGCAAUUAUCACUUAACUAUUGAGCC-------UUAUUCACUCAAAAUUGAAUAUCUA .........((((((((...(((.(((......))).)))...)))))(((((.((..(....)..))...))))).........)))-------.((((((........)))))).... ( -23.20) >consensus ACUACCAUCGGCGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCCAUUGUCACCAGACAAGUGAAUG_______UUAUACAUUCAAAACUAGAUAGUAA ............((.((((((....((.(((((.(((.....))).))))).))...))))))))....................................................... (-18.20 = -15.68 + -2.52)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:59:17 2006