| Sequence ID | sy_sa.0 |
|---|---|
| Location | 743,708 – 743,828 |
| Length | 120 |
| Max. P | 0.999996 |

| Location | 743,708 – 743,820 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -36.42 |
| Consensus MFE | -23.35 |
| Energy contribution | -19.38 |
| Covariance contribution | -3.97 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
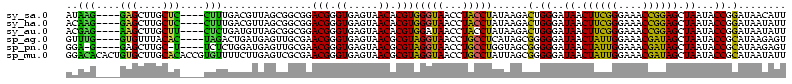
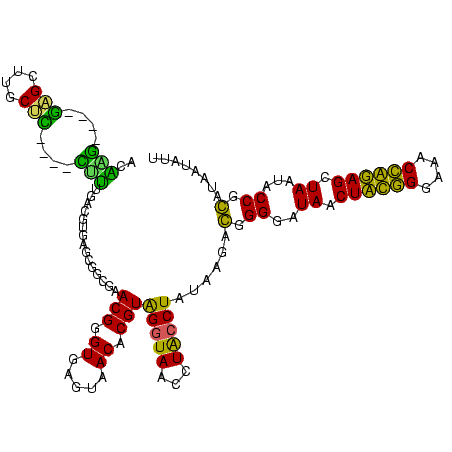
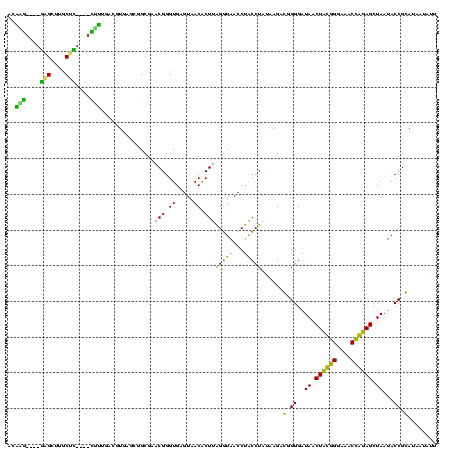
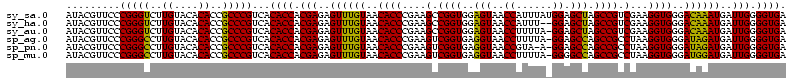
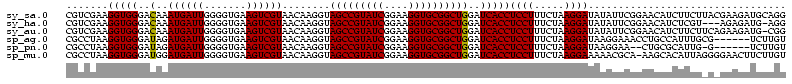
>sy_sa.0 743708 112 + 2516575/80-200 AUAAG----GAGCUUGCUC----CUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUU ....(----..(((..(((----.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))........ ( -37.30) >sy_ha.0 879828 112 + 2685015/80-200 ACAAG----GAGCUUGCUC----CUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUU ....(----..(((..(((----.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))........ ( -37.30) >sy_au.0 575711 112 - 2809422/80-200 ACGAG----AAGCUUGCUU----CUCUGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUU ..(((----(((....)))----))).(((((((.(((.((.((.(((.....))).)).....))..((((......))))....((((((....))))))......)))..))))))) ( -33.20) >sp_ag.0 16393 112 + 2160267/80-200 GUUUG----GUGUUUACAC----UAGACUGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGU (((((----(((....)))----)))))........(((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....)))))))....... ( -37.20) >sp_pn.0 243876 110 - 2038615/80-200 GGA-G----GAGCUUGC-U----UCUCUGGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUGGUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGU (((-(----(((....)-)----)))))........(((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....)))))))....... ( -36.70) >sp_mu.0 341842 120 - 2030921/80-200 GGACACACUGUGCUUGCACACCGUGUUUUCUUGAGUCGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUAUUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAUAUU ((((((..((((....))))..)))))).........((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....))))))........ ( -36.80) >consensus ACAAG____GAGCUUGCUC____CUUUGACGUGAGCGGCGAACGGGUGAGUAACACGUAGGUAACCUACCUAUAAGACGGGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAUAUU ..(((....(((....)))....)))...............(((.((.....)).)))(((((...)))))......(.((..((.((((((....)))))).))...)).)........ (-23.35 = -19.38 + -3.97)
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -32.67 |
| Energy contribution | -26.45 |
| Covariance contribution | -6.22 |
| Combinations/Pair | 1.73 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/120-240 GACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAU ........(((((.(((((((((...)))))))....((((..((.((((((....)))))).))...))))...((.(((((((((....)))))))))))........)).))))).. ( -37.20) >sy_ha.0 879828 120 + 2685015/120-240 GACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAU .....(((.....)))(((((((...)))))))....((((..((.((((((....)))))).))...))))........(((((((....)))))))(((..(((.....)))..))). ( -37.70) >sy_au.0 575711 120 - 2809422/120-240 GACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGU .(((((((.....)))((((.....))))...((((((((((.((.((((((....)))))).))...))........(((((((((....)))))))))........)))))))))))) ( -37.40) >sp_ag.0 16393 119 + 2160267/120-240 AACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGC-UU .(((.((.....)).)))((((((..(((..(...((((....((.((((((....)))))).))...))))....(((((((((((....)))))))))))....)..))).))))-)) ( -37.20) >sp_pn.0 243876 119 - 2038615/120-240 AACGGGUGAGUAACGCGUAGGUAACCUGCCUGGUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGC-AU .....(..(((........((((.(((((......)))))......((((((....))))))....))))((((..(((..((((((....))))))..))).....)))))))..)-.. ( -41.30) >sp_mu.0 341842 119 - 2030921/120-240 AACGGGUGAGUAACGCGUAGGUAACCUGCCUAUUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAUAUUAAUUAUUGCAUGAUAAUUGAUUGAAAGAUGCAAGCGC-AU ..............((((.((((.(((((......)))))......((((((....))))))....))))((((...((((((((((....))))))))))......))))..))))-.. ( -36.60) >consensus AACGGGUGAGUAACACGUAGGUAACCUACCUAUAAGACGGGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAUAUUUAGAACCGCAUGGUACUAAAGUGAAAGAUGCAAUUGC_AU .(((.((.....)).)))(((((...)))))......(.((..((.((((((....)))))).))...)).)......(((((((((....))))))))).................... (-32.67 = -26.45 + -6.22)
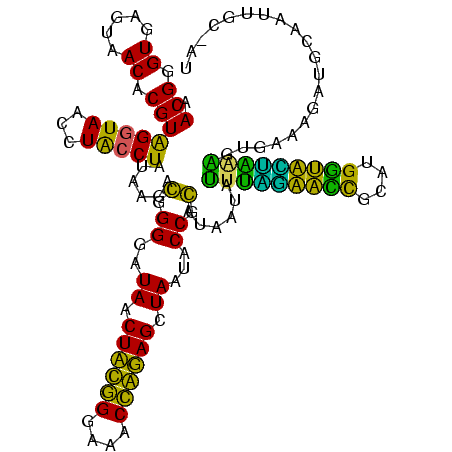
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -32.64 |
| Energy contribution | -26.70 |
| Covariance contribution | -5.94 |
| Combinations/Pair | 1.71 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/160-280 GGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAA ...(((((((((....))))(((((((((.((......(((((((((....)))))))))(((...(((((.....))))).((.......))...))).)))))))))))))))).... ( -39.20) >sy_ha.0 879828 120 + 2685015/160-280 GGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAA ...(((((((((....))))(((((((((((((......))))...(((..((((((((((..(((.....)))..))))).(.....)..))))).)))..)))))))))))))).... ( -39.90) >sy_au.0 575711 120 - 2809422/160-280 GGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAA ......((((((....))))))....((((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))..(((((....)).))).)))). ( -39.40) >sp_ag.0 16393 119 + 2160267/160-280 GGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGC-UUCACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGA ...(((((((((....))))(((((((((((((...(((((((((((....)))))))))))....(((((....))-)))...............))))..)))))))))))))).... ( -37.10) >sp_pn.0 243876 119 - 2038615/160-280 GGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGC-AUCACUACCAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGG ...(((((((((....))))(((((((((((((...(((..((((((....))))))..)))....(((((....))-)))....((....))...))))..)))))))))))))).... ( -40.20) >sp_mu.0 341842 119 - 2030921/160-280 GGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAUAUUAAUUAUUGCAUGAUAAUUGAUUGAAAGAUGCAAGCGC-AUCACUAGUAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUAA ...(((((((((....))))(((((((((.((((...((((((((((....))))))))))......)))).(((((-(...(((.....)))...)))))))))))))))))))).... ( -35.20) >consensus GGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAUAUUUAGAACCGCAUGGUACUAAAGUGAAAGAUGCAAUUGC_AUCACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUAA ...(((((((((....))))(((((((((.........(((((((((....)))))))))......((((......).))).....................)))))))))))))).... (-32.64 = -26.70 + -5.94)
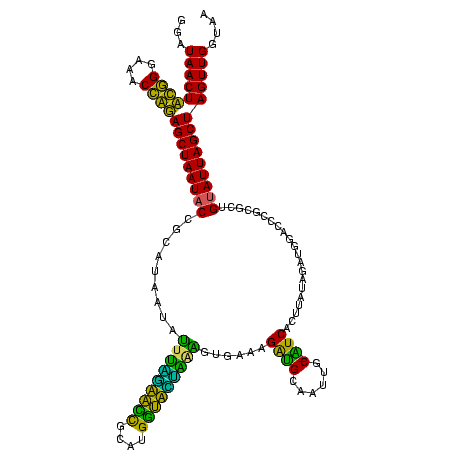
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -18.22 |
| Energy contribution | -16.70 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/160-280 UUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCC .....(((((((((((((.(.((((((...((((.((((((.((.........)).))))))))))))).)))(((..(....)..)))).))))))))).(((....))).)))).... ( -29.70) >sy_ha.0 879828 120 + 2685015/160-280 UUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCC .....(((((((((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......)))))))))))))).(((....))).)))).... ( -32.60) >sy_au.0 575711 120 - 2809422/160-280 UUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCC .....(((((((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....)))))))))))))).(((....))).)))).... ( -33.00) >sp_ag.0 16393 119 + 2160267/160-280 UCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGAA-GCAAUUGCUCCUUUUAAAUAACUAACAUGUGUUAAUUACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUCC ........((((((((((..((((((......))..((((((-((....))).........((((........)))).)))))....))))))))))))))................... ( -20.80) >sp_pn.0 243876 119 - 2038615/160-280 CCACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGUGAU-GCAAGUGCACCUUUUAAGCAAAUGUCAUGCAACAUCCACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUCC ........((((((((((..(((((((.(((...(((....)-))..))).)))......(((.......)))..............))))))))))))))................... ( -26.70) >sp_mu.0 341842 119 - 2030921/160-280 UUACCAACUAGCUAAUACAACGCAGGUCCAUCUACUAGUGAU-GCGCUUGCAUCUUUCAAUCAAUUAUCAUGCAAUAAUUAAUAUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUCC ........((((((((((...((((((.((((.......)))-).))))))...................((((.((((....))))))))))))))))))................... ( -29.60) >consensus UUACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGUGAU_GCAAAACCAUCUUUCAAUUAAGAACCAUGCGGUACCAAAUAUUAUCCGGUAUUAGCUACCGUUUCCAAAAGUUAUCC .........(((((((((...((.....(((......)))...)).................(((((((....)))))))...........))))))))).................... (-18.22 = -16.70 + -1.52) # Strand winner: forward (1.00)

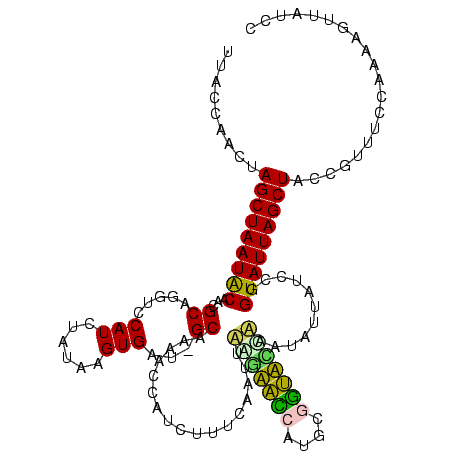
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.91 |
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -24.82 |
| Energy contribution | -22.30 |
| Covariance contribution | -2.52 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645309 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/200-320 UCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCA ..((((.((((.(((..(((.((.((((((((.......))))))))...))....)))..))))))).))))..((((((.((.........)).))))))..(((((....))))).. ( -31.80) >sy_ha.0 879828 120 + 2685015/200-320 UCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGA ..((((.((((.(((..(((.((.((((((((.......))))))))...))....)))..))))))).))))...(((((.((.........)).))))).(((((((....))))))) ( -35.20) >sy_au.0 575711 120 - 2809422/200-320 UCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAA ...((((.((..(((((....((.((((((((.......))))))))...))..)))))..)).)))).......((((((.(((......).)).))))))(((((((....))))))) ( -35.60) >sp_ag.0 16393 119 + 2160267/200-320 UCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGAA-GCAAUUGCUCCUUUUAAAUAACUAACAUGUGUUAAUU ..(((.((((........)))))))(((((((.......)))))))............((((((.((............(.(-((....))).).............)).)))))).... ( -23.91) >sp_pn.0 243876 119 - 2038615/200-320 UCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGUGAU-GCAAGUGCACCUUUUAAGCAAAUGUCAUGCAACAUCC ..((((..(((.(((((((..(((..((((..(((((........)))..((.........)).))..)))).)))..))))-))))))..)))).....(((.......)))....... ( -30.30) >sp_mu.0 341842 119 - 2030921/200-320 UCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCUCUUACCUUACCAACUAGCUAAUACAACGCAGGUCCAUCUACUAGUGAU-GCGCUUGCAUCUUUCAAUCAAUUAUCAUGCAAUAAUU ..(((.((((........)))))))(((((((.......)))))))...............((((((.((((.......)))-).))))))............................. ( -27.60) >consensus UCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGUGAU_GCAAAACCAUCUUUCAAUUAAGAACCAUGCGGUACCA ...((((.((..(((((....((.((((((((.......))))))))...))..)))))..)).)).)).................................(((((((....))))))) (-24.82 = -22.30 + -2.52) # Strand winner: forward (1.00)
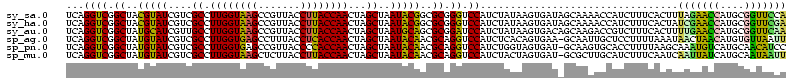
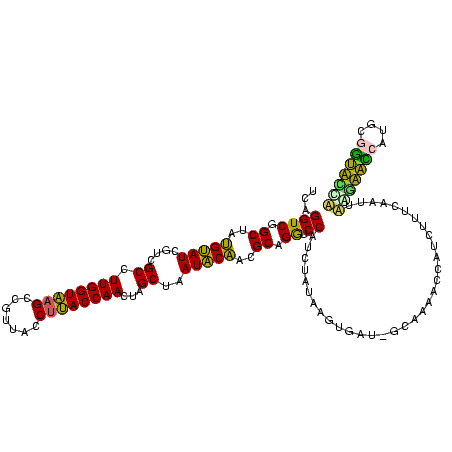
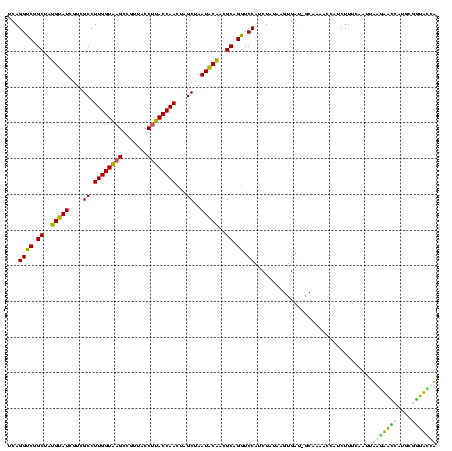
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -39.81 |
| Energy contribution | -40.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/240-360 CACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGA ..........((((((((((((((....))...((((((((.......)))))))))))).))....((.((((((((....)))...))))).))................)))))).. ( -41.90) >sy_ha.0 879828 120 + 2685015/240-360 CACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGA ..........((((((((((((((....))...((((((((.......)))))))))))).))....((.((((((((....)))...))))).))................)))))).. ( -41.90) >sy_au.0 575711 120 - 2809422/240-360 CACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGA ..........((((.(((.((((((((.(((..((((((((.......)))))))))))...)))))...((((((((....)))...))))).............)).).))))))).. ( -38.70) >sp_ag.0 16393 120 + 2160267/240-360 CACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGAGGUAAAGGCUCACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGA ...((((..............((((((.(((..((((((((.......)))))))))))...))))))..((((((((....)))...)))))))))(((((.(((.....)))))))). ( -44.50) >sp_pn.0 243876 120 - 2038615/240-360 CACUACCAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGA ....(((((......))).))((((((.(((..((((((((.......)))))))))))...))))))..((((((((....)))...)))))....(((((.(((.....)))))))). ( -42.10) >sp_mu.0 341842 120 - 2030921/240-360 CACUAGUAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUAAGGUAAGAGCUUACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGA .....((((......))))..((((((.(((..((((((((.......)))))))))))...))))))..((((((((....)))...)))))....(((((.(((.....)))))))). ( -42.50) >consensus CACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGCCCAGA ..........(((.((.((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....))))....)))))..(((((.(((.....)))))))). (-39.81 = -40.37 + 0.56)

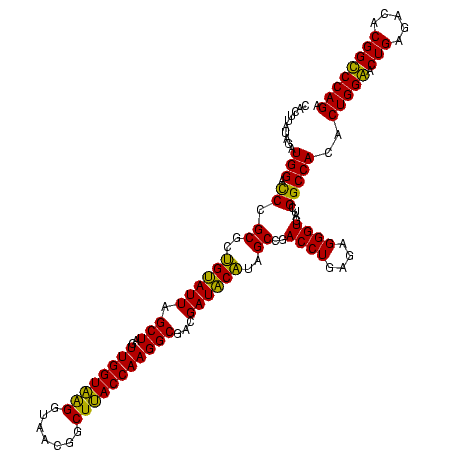
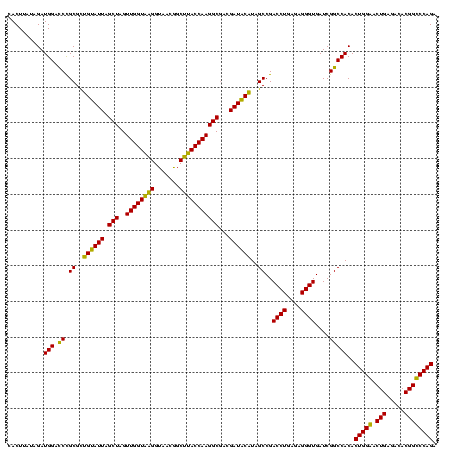
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -39.44 |
| Energy contribution | -38.33 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/240-360 UCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUG ..(((((((((((...........((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..))))))))).)))))).......... ( -43.90) >sy_ha.0 879828 120 + 2685015/240-360 UCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUG ..(((((((((((...........((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..))))))))).)))))).......... ( -43.90) >sy_au.0 575711 120 - 2809422/240-360 UCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUG ..((((((((((..((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...))..)))))).)))).......... ( -42.40) >sp_ag.0 16393 120 + 2160267/240-360 UCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUG ..((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).)))))).......... ( -39.80) >sp_pn.0 243876 120 - 2038615/240-360 UCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGUG ..((((((((((............((((((((((.........))))))))))((((....((.((((((...........))))))...))..)))).)))).)))))).......... ( -35.40) >sp_mu.0 341842 120 - 2030921/240-360 UCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCUCUUACCUUACCAACUAGCUAAUACAACGCAGGUCCAUCUACUAGUG ..((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).)))))).......... ( -37.50) >consensus UCUGGACCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGUG ..((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).)))))).......... (-39.44 = -38.33 + -1.11) # Strand winner: forward (0.56)
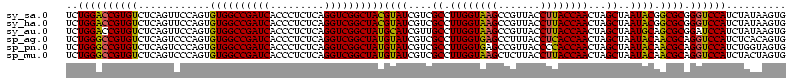
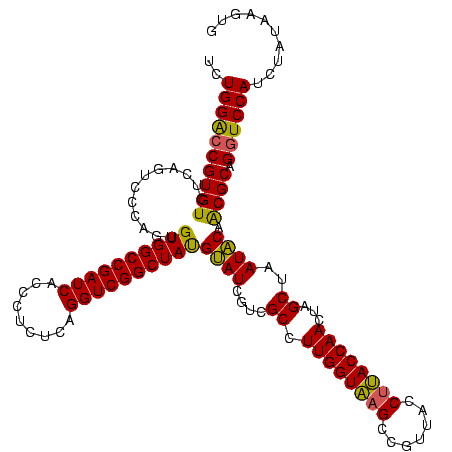
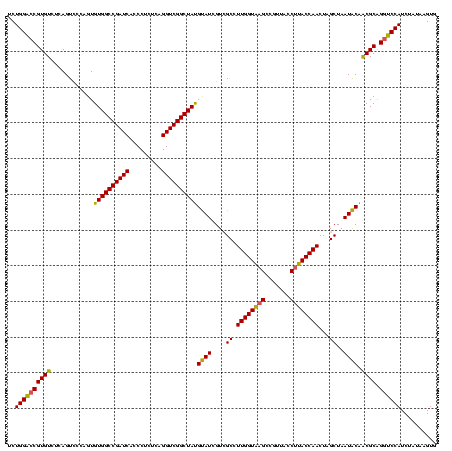
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.25 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -36.42 |
| Energy contribution | -32.15 |
| Covariance contribution | -4.27 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.80 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/600-720 CGCGUAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAU ((((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....)))....... ( -35.70) >sy_ha.0 879828 120 + 2685015/600-720 CGCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAU ((((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....)))....... ( -33.40) >sy_au.0 575711 120 - 2809422/600-720 CGCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAU ((((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....)))....... ( -33.40) >sp_ag.0 16393 119 + 2160267/600-720 AGCGCAGGCGGUUCUUUAAGUCUGAAGUUAAAGGCAGUGGCUUAACCAUUGUACG-CUUUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAU .(((((..((((((((((..((..((((.....(((((((.....)))))))..)-)))..))...)))))))).))..)))...........((((....)))).....))........ ( -35.80) >sp_pn.0 243876 119 - 2038615/600-720 AGCGCAGGCGGUUAGAUAAGUCUGAAGUUAAAGGCUGUGGCUUAACCAUAGUAGG-CUUUGGAAACUGUUUAACUUGAGUGCAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAU .(((((..((((((((((..((..(((((....(((((((.....))))))).))-)))..))...)))))))).))..)))...........((((....)))).....))........ ( -35.90) >sp_mu.0 341842 119 - 2030921/600-720 AGCGCAGGCGGUCAGGAAAGUCUGGAGUAAAAGGCUAUGGCUCAACCAUAGUGUG-CUCUGGAAACUGUCUGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAU .(((((..((((((((....((..(((((....(((((((.....))))))).))-)))..)).....)))))).))..)))...........((((....)))).....))........ ( -40.70) >consensus AGCGCAGGCGGUUUUUUAAGUCUGAAGUGAAAGCCCACGGCUCAACCAUGGAGGG_CAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAU .(((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....))........ (-36.42 = -32.15 + -4.27)
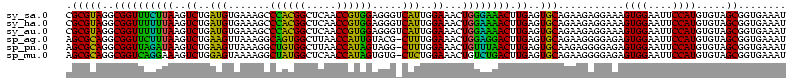
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -44.15 |
| Consensus MFE | -42.79 |
| Energy contribution | -41.23 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.42 |
| SVM RNA-class probability | 0.999986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/680-800 GCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGA (((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..)))))..((.(((((..........))))).)).. ( -42.10) >sy_ha.0 879828 120 + 2685015/680-800 GCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGA (((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..)))))..((.(((((..........))))).)).. ( -42.10) >sy_au.0 575711 120 - 2809422/680-800 GCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGA (((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..)))))..((.(((((..........))))).)).. ( -42.10) >sp_ag.0 16393 120 + 2160267/680-800 GCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCGAAAGCGUGGGGA (((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..)))))..((.(((((..........))))).)).. ( -46.30) >sp_pn.0 243876 120 - 2038615/680-800 GCAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGCUUGUAACUGACGCUGAGGCUCGAAAGCGUGGGGA (((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..)))))..((.(((((..........))))).)).. ( -45.50) >sp_mu.0 341842 120 - 2030921/680-800 GCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCAGUGGCGAAAGCGGCUCUCUGGUCUGUCACUGACGCUGAGGCUCGAAAGCGUGGGUA (((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..))))).(((.(((((..........))))).))). ( -46.80) >consensus GCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAAGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGA (((((...((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..)))))..((.(((((..........))))).)).. (-42.79 = -41.23 + -1.55)

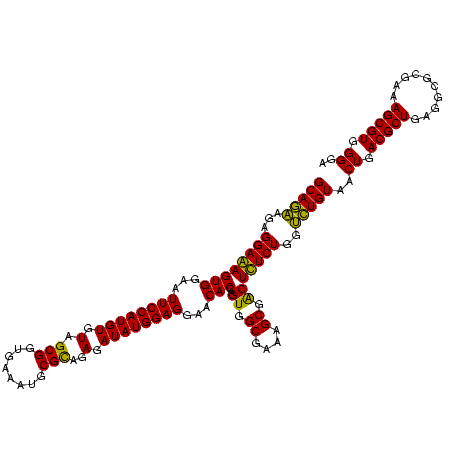
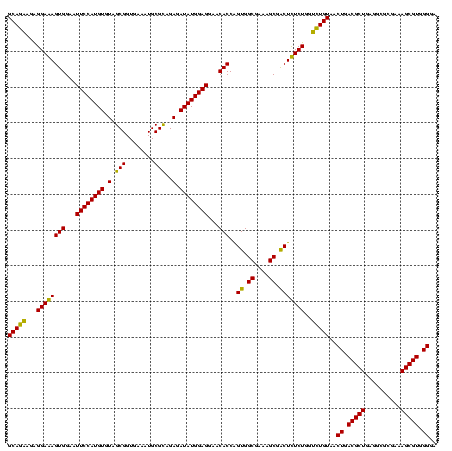
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -46.05 |
| Consensus MFE | -46.47 |
| Energy contribution | -46.05 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.33 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/800-920 UCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... ( -45.50) >sy_ha.0 879828 120 + 2685015/800-920 UCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... ( -45.50) >sy_au.0 575711 120 - 2809422/800-920 UCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... ( -45.50) >sp_ag.0 16393 120 + 2160267/800-920 GCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... ( -47.80) >sp_pn.0 243876 120 - 2038615/800-920 GCAAACAGGAUUAGAUACCCUGGUAGUCCACGCUGUAAACGAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... ( -44.20) >sp_mu.0 341842 120 - 2030921/800-920 GCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... ( -47.80) >consensus GCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGCCCUUUCCGCCCCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGAC .....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).)))... (-46.47 = -46.05 + -0.42)

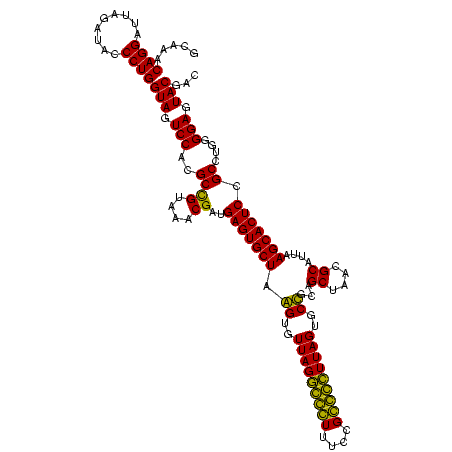
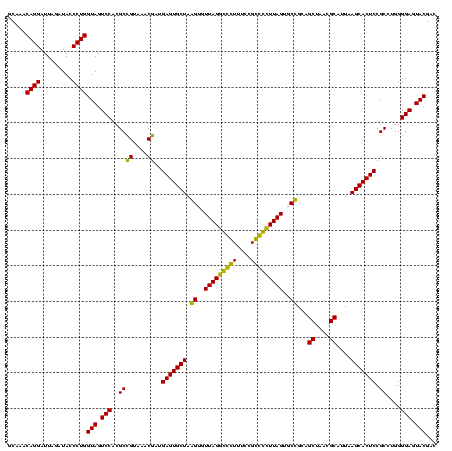
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -39.82 |
| Consensus MFE | -38.99 |
| Energy contribution | -37.38 |
| Covariance contribution | -1.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/800-920 GUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGA (((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).))))))...((((((.....))))))..... ( -42.40) >sy_ha.0 879828 120 + 2685015/800-920 GUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGA (((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).))))))...((((((.....))))))..... ( -42.40) >sy_au.0 575711 120 - 2809422/800-920 GUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGA (((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).))))))...((((((.....))))))..... ( -42.40) >sp_ag.0 16393 120 + 2160267/800-920 GUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGC (((.(((.((..((((((((((((....(.(((((..(((((.....)))((.....))))))))).)..)))))))..)))))..)).))))))...((((((.....))))))..... ( -38.90) >sp_pn.0 243876 120 - 2038615/800-920 GUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUACGGCACUAAACCCCGGAAAGGGUCUAACACCUAGCACUCAUCGUUUACAGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGC ..........(((.((.(((((((....(.((((((....)).....((((......)))).)))).)..)))))))...(....).)).))).....((((((.....))))))..... ( -34.90) >sp_mu.0 341842 120 - 2030921/800-920 GUCGUACUCCCCAGGCGGAGUGCUUAUUGCGUUAGCUCCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGC ((((((.(((...(((..((((((....((....))...))))))..)))..))).(((........))).............))))))((((((....(((((.....))))))))))) ( -37.90) >consensus GUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGCCCCGGAAACCCCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGA (((.(((.((..((((((((((((....(.((((((....)).....((((......)))).)))).)..)))))))..)))))..)).))))))...((((((.....))))))..... (-38.99 = -37.38 + -1.61) # Strand winner: forward (0.96)

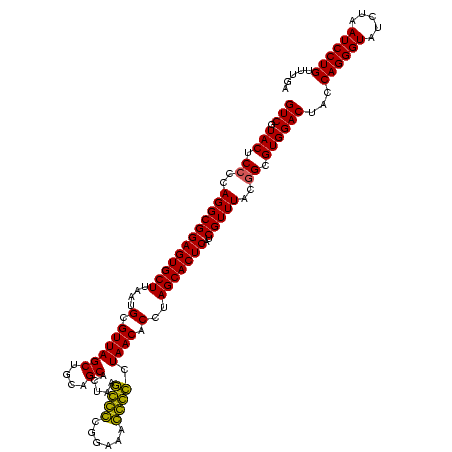
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -54.62 |
| Consensus MFE | -54.89 |
| Energy contribution | -54.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.92 |
| Structure conservation index | 1.01 |
| SVM decision value | 6.07 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/840-960 GAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). ( -53.80) >sy_ha.0 879828 120 + 2685015/840-960 GAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). ( -53.80) >sy_au.0 575711 120 - 2809422/840-960 GAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). ( -53.80) >sp_ag.0 16393 120 + 2160267/840-960 GAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). ( -56.10) >sp_pn.0 243876 120 - 2038615/840-960 GAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). ( -54.10) >sp_mu.0 341842 120 - 2030921/840-960 GAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). ( -56.10) >consensus GAUGAGUGCUAAGUGUUAGGCCCUUUCCGCCCCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCAC ...(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))). (-54.89 = -54.62 + -0.28)
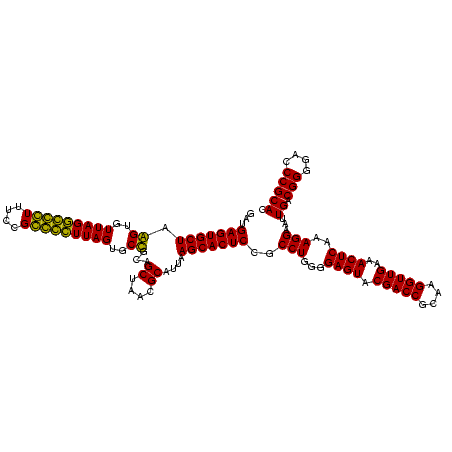
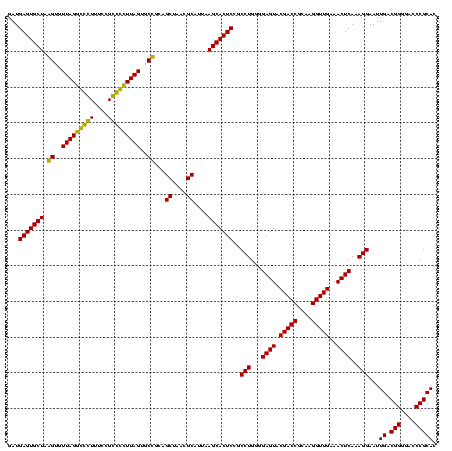
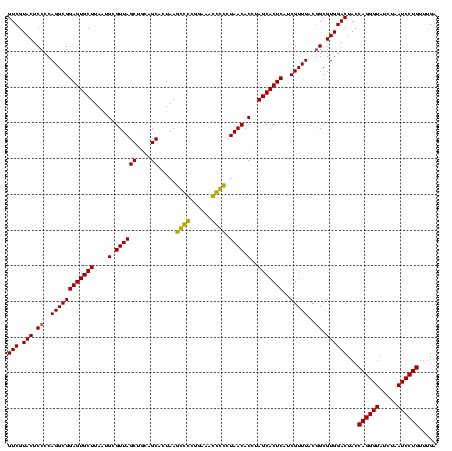
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -36.91 |
| Energy contribution | -35.13 |
| Covariance contribution | -1.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/840-960 GUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUC ..((((....)))).....(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))... ( -39.40) >sy_ha.0 879828 120 + 2685015/840-960 GUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUC ..((((....)))).....(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))... ( -39.40) >sy_au.0 575711 120 - 2809422/840-960 GUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUC ..((((....)))).....(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((.(....))))))))).)..)))))))... ( -39.40) >sp_ag.0 16393 120 + 2160267/840-960 GUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUC (((((((((((((...........((((..(.(((....))).).))))....(((..((((((....((....))...))))))..))).)))...))))))........))))..... ( -40.50) >sp_pn.0 243876 120 - 2038615/840-960 GUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUACGGCACUAAACCCCGGAAAGGGUCUAACACCUAGCACUCAUC ((((((....))((....((((((((((..(.(((....))).).)))).((.((.(.((((((....((....))...))))))...))).))))))))....)).....))))..... ( -35.50) >sp_mu.0 341842 120 - 2030921/840-960 GUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAUUGCGUUAGCUCCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUC (((((((((((((...........((((..(.(((....))).).))))....(((..((((((....((....))...))))))..))).)))...))))))........))))..... ( -40.50) >consensus GUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGCCCCGGAAACCCCCUAACACCUAGCACUCAUC ..((((....)))).....(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((......)))).)))).)..)))))))... (-36.91 = -35.13 + -1.78) # Strand winner: forward (0.98)

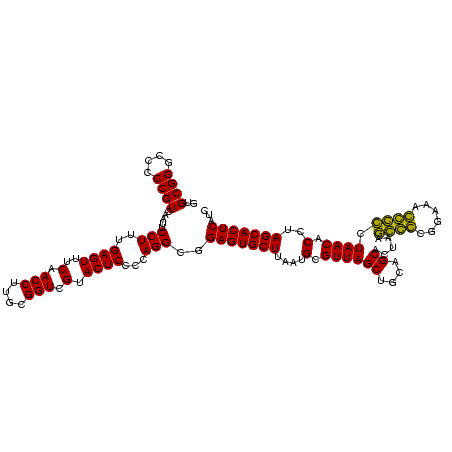
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.22 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -39.51 |
| Energy contribution | -39.48 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/880-1000 UGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.((....))(((((((.(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))......))))))).....)))........ ( -39.90) >sy_ha.0 879828 120 + 2685015/880-1000 UGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.((....))(((((((.(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))......))))))).....)))........ ( -39.90) >sy_au.0 575711 120 - 2809422/880-1000 UGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.((....))(((((((.(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))......))))))).....)))........ ( -39.90) >sp_ag.0 16393 120 + 2160267/880-1000 CGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.(((..((.(((((((.((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)).....))))))).)).)))...)))... ( -42.10) >sp_pn.0 243876 120 - 2038615/880-1000 CGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.(((..((.(((((((.((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)).....))))))).)).))).)))..... ( -41.80) >sp_mu.0 341842 120 - 2030921/880-1000 CGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.((....)).....((.((((((((...((((.(((((....)))))..))))..)))......))))))))))((((.((......)).))))......((((........)))). ( -39.70) >consensus CGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAG (((.(((..((.(((((((.((((((((...((((.(((((....)))))..))))..)))......))))).....((((.((......)).))))))))))).)).)))...)))... (-39.51 = -39.48 + -0.03)

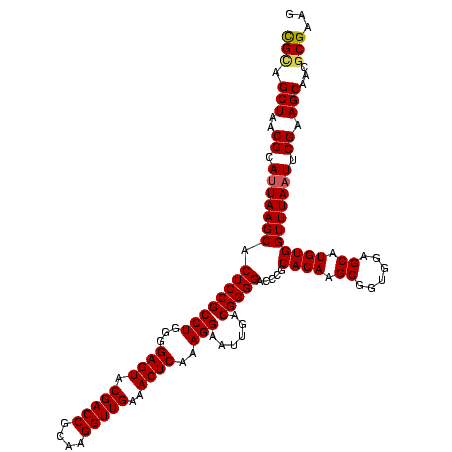
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -41.95 |
| Consensus MFE | -40.32 |
| Energy contribution | -41.60 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1080-1200 CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCG ...(((((((..(.(((((((((....)))((.(((......))))).))))))(((.......(((((((.((((..(.....)...))))..)))))))...))).)..))).)))). ( -42.50) >sy_ha.0 879828 120 + 2685015/1080-1200 CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCG ...(((((((..(.(((((((((....)))((.(((......))))).))))))(((.......(((((((.((((..(.....)...))))..)))))))...))).)..))).)))). ( -42.80) >sy_au.0 575711 120 - 2809422/1080-1200 CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCG ...(((((((..(.(((((((((....)))((.(((......))))).))))))(((.......(((((((.((((..(.....)...))))..)))))))...))).)..))).)))). ( -42.80) >sp_ag.0 16393 120 + 2160267/1080-1200 CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCG ...(((.(((....(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..)))))))))).)))(((.....))). ( -41.20) >sp_pn.0 243876 120 - 2038615/1080-1200 CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUCAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCG ...(((.(((....(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..)))))))))).)))(((.....))). ( -41.20) >sp_mu.0 341842 120 - 2030921/1080-1200 CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCG ...(((.(((....(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..)))))))))).)))(((.....))). ( -41.20) >consensus CAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAACAAACCG ...(((.(((....(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..)))))))))).)))(((.....))). (-40.32 = -41.60 + 1.28)

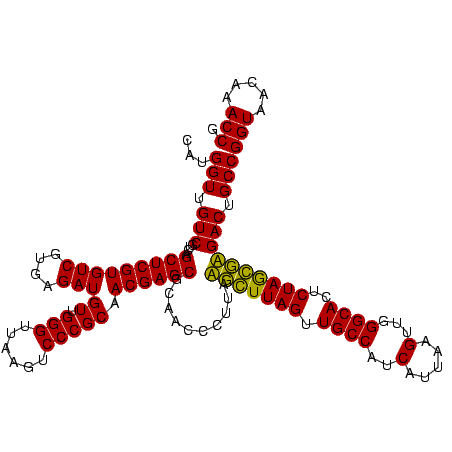
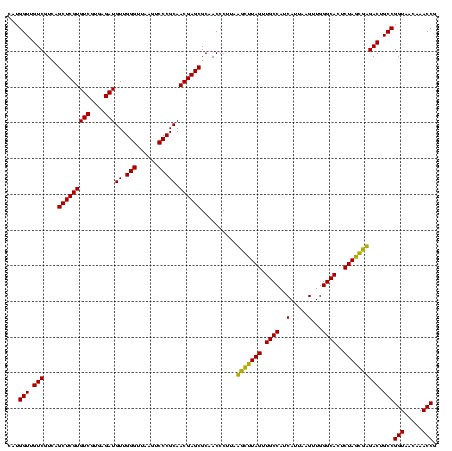
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -37.63 |
| Consensus MFE | -38.29 |
| Energy contribution | -36.90 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.92 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1120-1240 GUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ((..(((.....)))..))((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).... ( -38.20) >sy_ha.0 879828 120 + 2685015/1120-1240 GUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ((..(((.....)))..))((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).... ( -38.50) >sy_au.0 575711 120 - 2809422/1120-1240 GUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ((..(((.....)))..))((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).... ( -38.50) >sp_ag.0 16393 120 + 2160267/1120-1240 GUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ....(((.....)))..((((((.(((((((.((((..(.....)...))))..))))))).....(((((.....))))))))..)))((((((((.(......).)))).)))).... ( -36.60) >sp_pn.0 243876 120 - 2038615/1120-1240 GUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUCAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ....(((.....)))..((((((.(((((((.((((..(.....)...))))..))))))).....(((((.....))))))))..)))((((((((.(......).)))).)))).... ( -36.60) >sp_mu.0 341842 120 - 2030921/1120-1240 GUCCCGCAACGAGCGCAACCCUUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ((..(((.....)))..))((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).... ( -37.40) >consensus GUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAU ((..(((.....)))..))((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).... (-38.29 = -36.90 + -1.39)

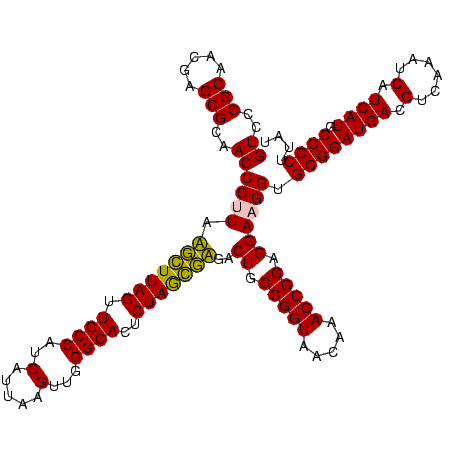

| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -38.68 |
| Energy contribution | -38.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1120-1240 AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))....(((((((((.....)))))..(.((((...((((((....(((((.(..(....)....))))))....))))))))))))))). ( -39.70) >sy_ha.0 879828 120 + 2685015/1120-1240 AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))....(((((((((.....)))))..(.((((...((((((....(((((.(..(....)....))))))....))))))))))))))). ( -39.70) >sy_au.0 575711 120 - 2809422/1120-1240 AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))....(((((((((.....)))))..(.((((...((((((....(((((.(..(....)....))))))....))))))))))))))). ( -39.70) >sp_ag.0 16393 120 + 2160267/1120-1240 AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAGGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))........(((((.....)))))..(((((((..((((((....((((.((..(....)...)).))))....))))))...))))))) ( -47.80) >sp_pn.0 243876 120 - 2038615/1120-1240 AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUGAAUGAUGGCAACUAACAAUAGGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))........(((((.....)))))..(((((((..((((((.(..(((......(....)......)))..)..))))))...))))))) ( -46.00) >sp_mu.0 341842 120 - 2030921/1120-1240 AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAAGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))........(((((.....)))))..(((((((..((((((....((((.((..(....)...)).))))....))))))...))))))) ( -48.10) >consensus AUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAAGGGUUGCGCUCGUUGCGGGAC ....((((.((((((......))))))))))....((((.((((.....)))((((.((......)).))))......(((((..((((((.........))))))..)))))).)))). (-38.68 = -38.68 + -0.00) # Strand winner: reverse (0.72)

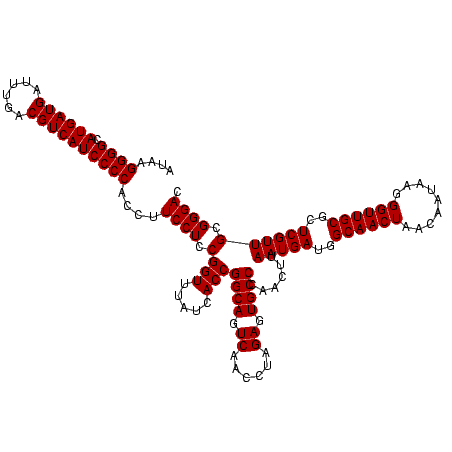
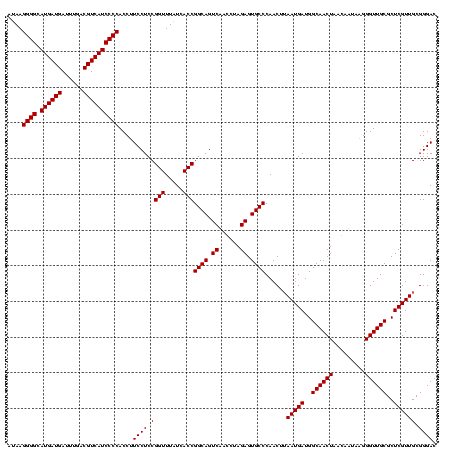
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -37.36 |
| Energy contribution | -34.28 |
| Covariance contribution | -3.08 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1240-1360 GAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCUAAACCGCGAGGUCAUGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAG ((((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..))))))))))((((((........))))))...... ( -31.90) >sy_ha.0 879828 120 + 2685015/1240-1360 GAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAG ((((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..))))))))))((((((........))))))...... ( -32.30) >sy_au.0 575711 120 - 2809422/1240-1360 GAUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAG ((((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..))))))))))((((((........))))))...... ( -32.30) >sp_ag.0 16393 120 + 2160267/1240-1360 GACCUGGGCUACACACGUGCUACAAUGGUUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAAUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAG ((.((((((.((....))))).....(((((((.....(((..((..((((....))))..))....))).....))))))).))).))....(((((((........)))))))..... ( -39.90) >sp_pn.0 243876 120 - 2038615/1240-1360 GACCUGGGCUACACACGUGCUACAAUGGCUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAGUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAG ((.((((((.((....))))).....(((((((.....(((..((..((((....))))..))....))).....))))))).))).))....(((((((........)))))))..... ( -42.20) >sp_mu.0 341842 120 - 2030921/1240-1360 GACCUGGGCUACACACGUGCUACAAUGGUCGGUACAACGAGUUGCGAGCCGGUGACGGCAAGCUAAUCUCUGAAAGCCGAUCUCAGUUCGGAUUGGAGGCUGCAACUCGCCUCCAUGAAG ((((..(((.((....))))).....))))((.....((((((((..((((....)))).(((....(((..(...((((.......)))).)..))))))))))))))...))...... ( -46.40) >consensus GACCUGGGCUACACACGUGCUACAAUGGACAAUACAAAGAGCAGCGAAACCGCGACGGCAAGCAAAUCCCAUAAAGCCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGACUACAUGAAG ((.((((((.((....))))).....(((((((.....(((..((...(((....)))...))....))).....))))))).))).))....(((((((........)))))))..... (-37.36 = -34.28 + -3.08)
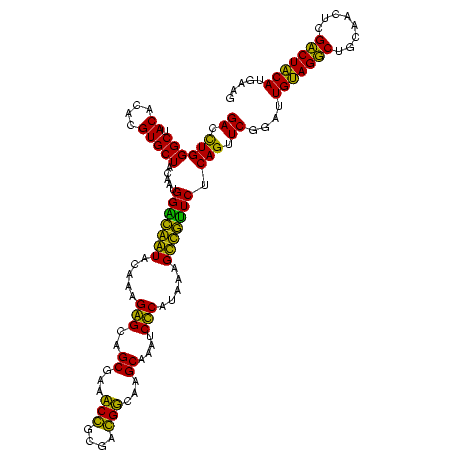
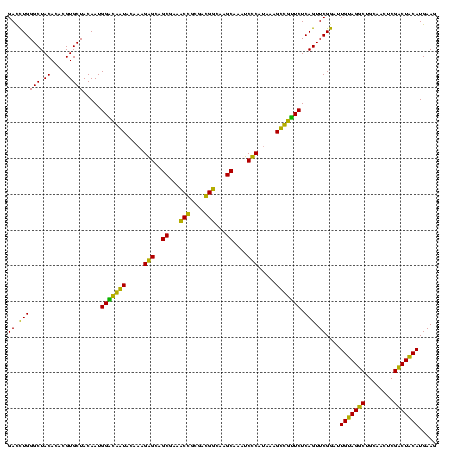
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -38.29 |
| Energy contribution | -36.27 |
| Covariance contribution | -2.03 |
| Combinations/Pair | 1.15 |
| Mean z-score | -0.74 |
| Structure conservation index | 1.06 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1320-1440 UCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGCUAGUAAUCGUAGAUCAGCAUGCUACGGUGAAUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. ( -32.20) >sy_ha.0 879828 120 + 2685015/1320-1440 UCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGCUAGUAAUCGUAGAUCAGCAUGCUACGGUGAAUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. ( -32.20) >sy_au.0 575711 120 - 2809422/1320-1440 UCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGCUAGUAAUCGUAGAUCAGCAUGCUACGGUGAAUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. ( -32.20) >sp_ag.0 16393 120 + 2160267/1320-1440 UCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGUCGGAAUCGCUAGUAAUCGCGGAUCAGCACGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. ( -39.60) >sp_pn.0 243876 120 - 2038615/1320-1440 UCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGUCGGAAUCGCUAGUAAUCGCGGAUCAGCACGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. ( -39.60) >sp_mu.0 341842 120 - 2030921/1320-1440 UCUCAGUUCGGAUUGGAGGCUGCAACUCGCCUCCAUGAAGUCGGAAUCGCUAGUAAUCGCGGAUCAGCACGCCGCGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. ( -41.80) >consensus UCUCAGUUCGGAUUGUAGGCUGCAACUCGACUACAUGAAGCCGGAAUCGCUAGUAAUCGCAGAUCAGCACGCCACGGUGAAUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCAC .........((..(((((((........)))))))(((....((((.((.((...(((((((.........)))))))...))))))))(((((..((....))..))))))))..)).. (-38.29 = -36.27 + -2.03)

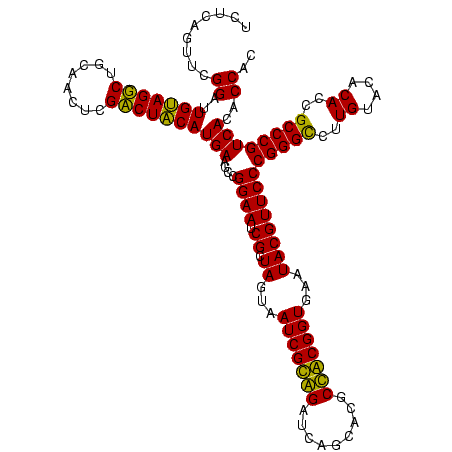
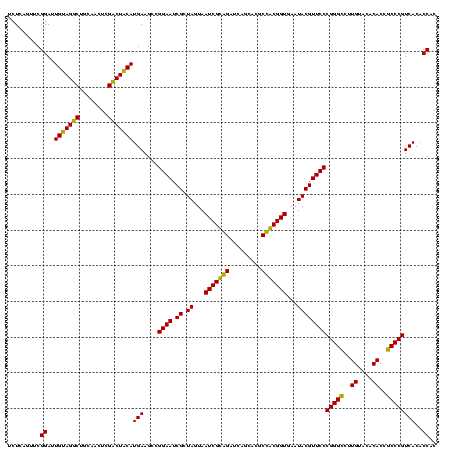
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -41.17 |
| Energy contribution | -39.78 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1400-1520 AUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..((((((..(((((((...((((..(((..(((....))).))).)))))))..))))..))))))..))).)))). ( -41.40) >sy_ha.0 879828 118 + 2685015/1400-1520 AUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUU--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..((((((..(((((((...((((..(((..((....--)).))).)))))))..))))..))))))..))).)))). ( -40.80) >sy_au.0 575711 119 - 2809422/1400-1520 AUACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..((((((..(((((((...((((..(((..((.....-)).))).)))))))..))))..))))))..))).)))). ( -40.70) >sp_ag.0 16393 119 + 2160267/1400-1520 AUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((.....-)).))).)))).))..))))..))))))..))).)))). ( -42.10) >sp_pn.0 243876 118 - 2038615/1400-1520 AUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCGUA-A-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..((((((..((((...((.((((..(((..((...-.-)).))).)))).))..))))..))))))..))).)))). ( -44.60) >sp_mu.0 341842 119 - 2030921/1400-1520 AUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGGGCCAGCCGCCUAAGGUGGGAUGGAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..(((..(..((((...((.((((..(((..(((....-)))))).)))).))..))))..)..)))..))).)))). ( -42.50) >consensus AUACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGAAGUAACCUUUUA_GGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGA .........(((((..((....))..)))))...((((.(((..((((((..((((....(.((((..(((..((......)).))).)))).)...))))..))))))..))).)))). (-41.17 = -39.78 + -1.39)
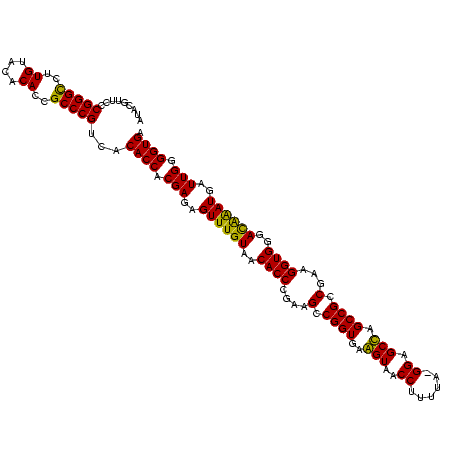

| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -41.67 |
| Energy contribution | -40.53 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.13 |
| SVM RNA-class probability | 0.999975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1440-1560 GAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....((((((..(((((((...((((..(((..(((....))).))).)))))))..))))..))))))((((.......)))).........((((((((((....))))))))))... ( -42.90) >sy_ha.0 879828 118 + 2685015/1440-1560 GAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUU--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....((((((..(((((((...((((..(((..((....--)).))).)))))))..))))..))))))((((.......)))).........((((((((((....))))))))))... ( -42.30) >sy_au.0 575711 119 - 2809422/1440-1560 GAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....((((((..(((((((...((((..(((..((.....-)).))).)))))))..))))..))))))((((.......)))).........((((((((((....))))))))))... ( -42.20) >sp_ag.0 16393 119 + 2160267/1440-1560 GAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....((((((..((((...((.((((..(((..((.....-)).))).)))).))..))))..))))))((((.......)))).........((((((((((....))))))))))... ( -42.10) >sp_pn.0 243876 118 - 2038615/1440-1560 GAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCGUA-A-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....((((((..((((...((.((((..(((..((...-.-)).))).)))).))..))))..))))))((((.......)))).........((((((((((....))))))))))... ( -44.60) >sp_mu.0 341842 119 - 2030921/1440-1560 GAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGGGCCAGCCGCCUAAGGUGGGAUGGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....(((((...(((((..(((((((......))).....-....(((.(((((...)))))..)))..)))).)))))....)).)))....((((((((((....))))))))))... ( -44.30) >consensus GAGAGUUUGUAACACCCGAAGCCGGUGAAGUAACCUUUUA_GGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAU ....((((((..((((....(.((((..(((..((......)).))).)))).)...))))..))))))((((.......)))).........((((((((((....))))))))))... (-41.67 = -40.53 + -1.14)

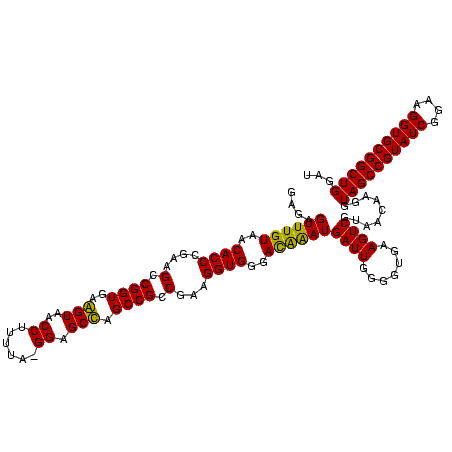
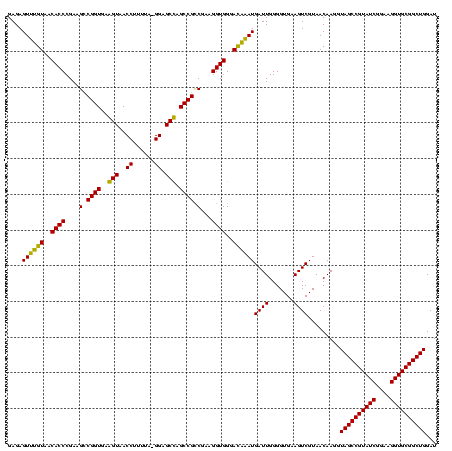
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -40.28 |
| Consensus MFE | -35.15 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1480-1600 UGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACG .((......))...(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((.....)))...)))))))..... ( -39.70) >sy_ha.0 879828 116 + 2685015/1480-1600 -GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---A -.(((....(((.....(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))......))).....)))..---. ( -38.40) >sy_au.0 575711 119 - 2809422/1480-1600 -GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAG -((......))(..(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((.....)))...)))))))..).. ( -39.80) >sp_ag.0 16393 117 + 2160267/1480-1600 -GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAAACCUGCCAUUUGCG-- -...((...(((((...)))))...(((((...(((((((.((....))....((((((((((....))))))))))..)))))(((((((....)).)))))..))...))))))).-- ( -41.90) >sp_pn.0 243876 114 - 2038615/1480-1600 -GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAA--CUGCGCAUUG-G-- -...((((((((((...)))))..((((.....(((((((.((....))....((((((((((....))))))))))..)))))))...))))...........--))).))....-.-- ( -41.70) >sp_mu.0 341842 118 - 2030921/1480-1600 -GGGGCCAGCCGCCUAAGGUGGGAUGGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAAAAACGCA-AAGCACAUUAGGGG -(.(.(((.(((((...)))))..)))..((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..........).).-.............. ( -40.20) >consensus _GGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAAUUCGGAACAUCUUUU_G_G .....((.(((......))).))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))............................. (-35.15 = -34.90 + -0.25)

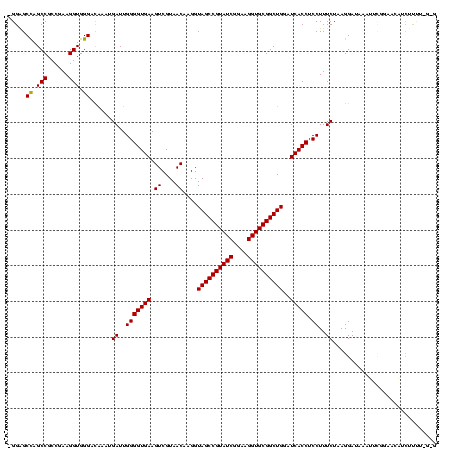
| Location | 743,708 – 743,828 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -31.18 |
| Energy contribution | -30.93 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913522 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 743708 120 + 2516575/1490-1610 CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGG ..((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))..(((((((....))))))).... ( -43.60) >sy_ha.0 879828 116 + 2685015/1490-1610 CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---AGAGAUG-AGG ..((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))..((((((..---.))))))-... ( -41.90) >sy_au.0 575711 119 - 2809422/1490-1610 CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAGAAGAUG-CGG ..((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))..(((((((....)))))))-... ( -43.90) >sp_ag.0 16393 114 + 2160267/1490-1610 CGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAAACCUGCCAUUUGCG------UCUUGU (((..(.((..((............(((((.((....))....((((((((((....))))))))))..)))))(((((((....)).)))))..))..)).)..)))------...... ( -41.50) >sp_pn.0 243876 111 - 2038615/1490-1610 CGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAGGAA--CUGCGCAUUG-G------UCUUGU ((((...))))((((.((((...(((((((.((....))....((((((((((....))))))))))..)))))(((((((....)).))))).--))...)))).-)------)))... ( -38.60) >sp_mu.0 341842 119 - 2030921/1490-1610 CGCCUAAGGUGGGAUGGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAAAAACGCA-AAGCACAUUAGGGGAACUUCUUGU (.(((((.(((...((((.....(((((((.((....))....((((((((((....))))))))))..)))))))...)))).(......)...-...))).))))).).......... ( -39.20) >consensus CGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAAAUUCGGAACAUCUUUU_G_GAAGAUCUUGG .......(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))................................. (-31.18 = -30.93 + -0.25)
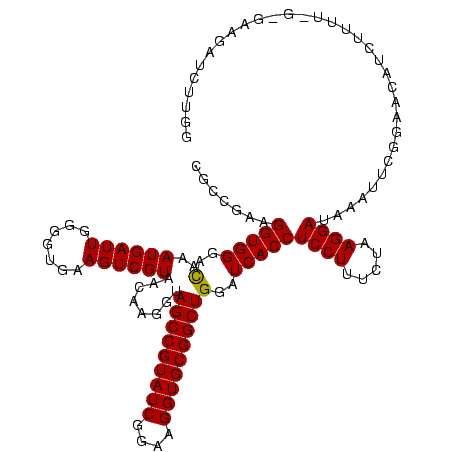
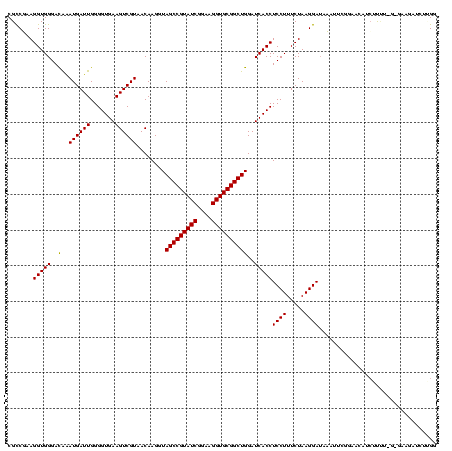
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:59:09 2006