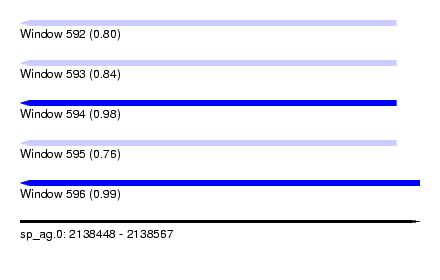
| Sequence ID | sp_ag.0 |
|---|---|
| Location | 2,138,448 – 2,138,567 |
| Length | 119 |
| Max. P | 0.994183 |

| Location | 2,138,448 – 2,138,560 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.56 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -16.84 |
| Energy contribution | -18.60 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 2138448 112 - 2160267/80-200 UAUAAAAUUGUAAUGCCGGCUACAUGACUUGAACACGCGACCCUCUGAUUACAAAUCAGAUGCUCUACCAACUGAGCUAAGCCGGCA-AUCUACUA-------AUGCGGGUGAAGGGACU .............((((((((......................((((((.....)))))).((((........))))..))))))))-(((..(..-------..)..)))......... ( -28.40) >sp_mu.0 1683404 104 + 2030921/80-200 ------------AUGCCGGCUACAUGACUUGAACACGCGACCCUCUGAUUACAAAUCAGAUGCUCUACCAACUGAGCUAAGCCGGCCUAUCUCCUCUAU----AUGCGGGUGAAGGGACU ------------..(((((((......................((((((.....)))))).((((........))))..)))))))((.((.(((....----....))).)).)).... ( -29.70) >sp_pn.0 1789331 120 + 2038615/80-200 UUCUAUCUAAAAAUGCCGGCUACAUGACUUGAACACGCGACCCUCUGAUUACAAAUCAGAUGCUCUACCAACUGAGCUAAGCCGGCUCAUUUGUUAUAUCUUAAUGCGGGUUAAGGGACU .....(((......(((((((......................((((((.....)))))).((((........))))..))))))).............((((((....))))))))).. ( -29.00) >sy_ha.0 2500854 110 + 2685015/80-200 UAAAACAAUCGUAUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAA--UGGCUCA-AAGAUG-------GUGCCGGCCAGAGGACU .......(((...(((((((((((((.......))))).....((((((.....)))))).((((........))))..)--)))))))-..)))(-------((....)))........ ( -32.90) >sy_sa.0 2312029 111 + 2516575/80-200 AAAUGAAACUGGAUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAA--UGGCUCAUGAAAUG-------GUGCCGGCCAGAGGACU ........((((((((((((((((((.......))))).....((((((.....)))))).((((........))))..)--))))))))....((-------....)).))))...... ( -36.50) >consensus UAAAAAAAU_GAAUGCCGGCUACAUGACUUGAACACGCGACCCUCUGAUUACAAAUCAGAUGCUCUACCAACUGAGCUAAGCCGGCUCAUCUAAUA_______AUGCGGGUCAAGGGACU ..............(((((((......................((((((.....)))))).((((........))))..))))))).................................. (-16.84 = -18.60 + 1.76)


| Location | 2,138,448 – 2,138,560 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.56 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -21.50 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 2138448 112 - 2160267/80-200 AGUCCCUUCACCCGCAU-------UAGUAGAU-UGCCGGCUUAGCUCAGUUGGUAGAGCAUCUGAUUUGUAAUCAGAGGGUCGCGUGUUCAAGUCAUGUAGCCGGCAUUACAAUUUUAUA .................-------..((((..-((((((((..((((........)))).((((((.....)))))).....(((((.......)))))))))))))))))......... ( -31.60) >sp_mu.0 1683404 104 + 2030921/80-200 AGUCCCUUCACCCGCAU----AUAGAGGAGAUAGGCCGGCUUAGCUCAGUUGGUAGAGCAUCUGAUUUGUAAUCAGAGGGUCGCGUGUUCAAGUCAUGUAGCCGGCAU------------ .(((((((.........----...)))).)))..(((((((..((((........)))).((((((.....)))))).....(((((.......))))))))))))..------------ ( -33.00) >sp_pn.0 1789331 120 + 2038615/80-200 AGUCCCUUAACCCGCAUUAAGAUAUAACAAAUGAGCCGGCUUAGCUCAGUUGGUAGAGCAUCUGAUUUGUAAUCAGAGGGUCGCGUGUUCAAGUCAUGUAGCCGGCAUUUUUAGAUAGAA .(((.(((((......))))).............(((((((..((((........)))).((((((.....)))))).....(((((.......)))))))))))).......))).... ( -31.20) >sy_ha.0 2500854 110 + 2685015/80-200 AGUCCUCUGGCCGGCAC-------CAUCUU-UGAGCCA--UUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCAUACGAUUGUUUUA ((((...((((((..((-------((...(-(((((..--...)))))).)))).((((.((((((((((.....)))))))))).))))..........)))).))...))))...... ( -35.90) >sy_sa.0 2312029 111 + 2516575/80-200 AGUCCUCUGGCCGGCAC-------CAUUUCAUGAGCCA--UUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGCCCACUAUGGCUCAUCCAGUUUCAUUU ......((((.....((-------((.....(((((..--...)))))..)))).((((..(((((((((.....)))))))))..))))(((((......)))))..))))........ ( -37.20) >consensus AGUCCCUUCACCCGCAU_______CAGCAGAUGAGCCGGCUUAGCUCAGUUGGUAGAGCAUCUGAUUUGUAAUCAGAGGGUCGCGUGUUCAAGUCAUGUAGCCGGCAUUC_AAUUUAUUA ..................................(((((((..((((........)))).((((((.....)))))).....(((((.......)))))))))))).............. (-21.50 = -22.26 + 0.76) # Strand winner: reverse (0.99)

| Location | 2,138,448 – 2,138,560 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.65 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 2138448 112 - 2160267/160-280 GCCGGCA-AUCUACUA-------AUGCGGGUGAAGGGACUUGAACCCCCACGCCGUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUUUCUAAAUGACC ...((..-((.((..(-------(((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........))))))))))..)).))..)) ( -39.10) >sp_mu.0 1683404 116 + 2030921/160-280 GCCGGCCUAUCUCCUCUAU----AUGCGGGUGAAGGGACUUGAACCCCCACGCCCUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUCUCUUGAUGACC ...((..((((........----((((((((((((((.(.((......)).))))))..(((((((((.....))))))))).................))))))))).....)))).)) ( -39.92) >sp_pn.0 1789331 116 + 2038615/160-280 GCCGGCUCAUUUGUUAUAUCUUAAUGCGGGUUAAGGGACUUGAACCCCCACGCCGUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCAAACCCGCAUAU----AUGACC ............(((((((....(((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........)))))))))))----))))). ( -38.20) >sy_ha.0 2500854 109 + 2685015/160-280 --UGGCUCA-AAGAUG-------GUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCUAA-UUAAUUAAGAAUGGUGGAG --.......-..(((.-------..(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))..)-))................. ( -30.00) >sy_sa.0 2312029 109 + 2516575/160-280 --UGGCUCAUGAAAUG-------GUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAGUUGAGCUAGGCCGGCUA--UUCAUUUGAAAUGGUGGAG --(.((.(((.(((((-------(.(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))..--.))))))...))))).).. ( -35.70) >consensus GCCGGCUCAUCUAAUA_______AUGCGGGUCAAGGGACUUGAACCCCCACGCCAUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCAAACCCGCAUAUAAUGAUGACC .........................(((((.(((.((...((......))..)).))).(((((((((.....)))))))))..........)))))....................... (-17.60 = -17.40 + -0.20)

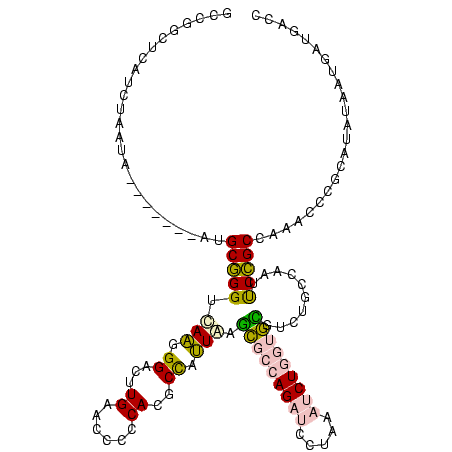

| Location | 2,138,448 – 2,138,560 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.65 |
| Mean single sequence MFE | -39.28 |
| Consensus MFE | -17.44 |
| Energy contribution | -16.24 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.758694 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 2138448 112 - 2160267/160-280 GGUCAUUUAGAAAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGAUUUAGGAUCUGGCGCUUAACGGCGUGGGGGUUCAAGUCCCUUCACCCGCAU-------UAGUAGAU-UGCCGGC (((.(((((..((((((((((.((.(((.(((((...(((.(((((((...))))))).(((....))))))...).)))).))))).)))))))))-------)..)))))-.)))... ( -47.00) >sp_mu.0 1683404 116 + 2030921/160-280 GGUCAUCAAGAGAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGAUUUAGGAUCUGGCGCUUAAGGGCGUGGGGGUUCAAGUCCCUUCACCCGCAU----AUAGAGGAGAUAGGCCGGC (((((((.....(((((((((.((.(((.(((((...(((.(((((((...))))))).(((....))))))...).)))).))))).)))))))))----........))).))))... ( -45.52) >sp_pn.0 1789331 116 + 2038615/160-280 GGUCAU----AUAUGCGGGUUUGGCGGAAUUGGCAGACGCACCAGAUUUAGGAUCUGGCGCUUAACGGCGUGGGGGUUCAAGUCCCUUAACCCGCAUUAAGAUAUAACAAAUGAGCCGGC ((((((----..(((((((((.((.(((.(((((...(((.(((((((...))))))).(((....))))))...).)))).))))).))))))))).............)))).))... ( -38.66) >sy_ha.0 2500854 109 + 2685015/160-280 CUCCACCAUUCUUAAUUAA-UUAGCCGGCCUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCAC-------CAUCUU-UGAGCCA-- ..........(((((....-...(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))..-------.....)-))))...-- ( -32.44) >sy_sa.0 2312029 109 + 2516575/160-280 CUCCACCAUUUCAAAUGAA--UAGCCGGCCUAGCUCAACUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCAC-------CAUUUCAUGAGCCA-- ..............(((((--..(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))..-------...)))))......-- ( -32.80) >consensus GGUCAUCAAGAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGAUUUAGGAUCUGGCGCUUAACGGCGUGGGGGUUCAAGUCCCUUCACCCGCAU_______CAGCAGAUGAGCCGGC .......................((((.........((((.((((((.....))))))((.....))))))(((((.......)))))...))))......................... (-17.44 = -16.24 + -1.20) # Strand winner: reverse (0.75)

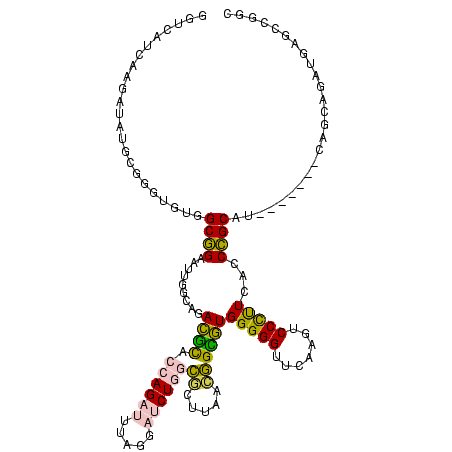
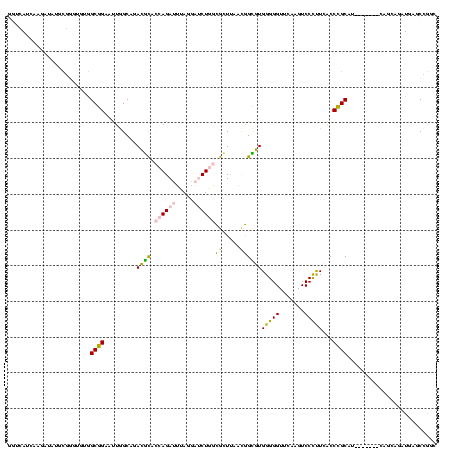
| Location | 2,138,448 – 2,138,567 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.80 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -24.46 |
| Energy contribution | -23.70 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_ag.0 2138448 119 - 2160267/182-302 -AUGCGGGUGAAGGGACUUGAACCCCCACGCCGUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUUUCUAAAUGACCCGUACUGGGCUCGAACCAGUGA -(((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........)))))))))..............((((((.......)))))). ( -43.60) >sp_mu.0 1683404 119 + 2030921/182-302 -AUGCGGGUGAAGGGACUUGAACCCCCACGCCCUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUCUCUUGAUGACCCGUACUGGGCUCGAACCAGUGA -((((((((((((((.(.((......)).))))))..(((((((((.....))))))))).................)))))))))..............((((((.......)))))). ( -45.90) >sp_pn.0 1789331 116 + 2038615/182-302 AAUGCGGGUUAAGGGACUUGAACCCCCACGCCGUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCAAACCCGCAUAU----AUGACCCGUACUGGGCUCGAACCAGUGA .(((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........)))))))))..----........((((((.......)))))). ( -42.20) >sy_ha.0 2500854 118 + 2685015/182-302 -GUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCUAA-UUAAUUAAGAAUGGUGGAGAAUGACGGGUUCGAACCGCCGA -..(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))...-...........((((((.((((.....))))...)))))). ( -39.80) >sy_sa.0 2312029 117 + 2516575/182-302 -GUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAGUUGAGCUAGGCCGGCUA--UUCAUUUGAAAUGGUGGAGGAUGACGGGUUCGAACCGCCGA -....((((....((..((((((((.......((((((.....))))))..((((((((((((.((...))))))).--(((....)))..)))))))......)))))))).)))))). ( -39.50) >consensus _AUGCGGGUCAAGGGACUUGAACCCCCACGCCAUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCAAACCCGCAUAUAAUGAUGACCCGUACUGGGCUCGAACCAGUGA ...(((((.(((.((...((......))..)).))).(((((((((.....)))))))))..........))))).........................((((((.......)))))). (-24.46 = -23.70 + -0.76)

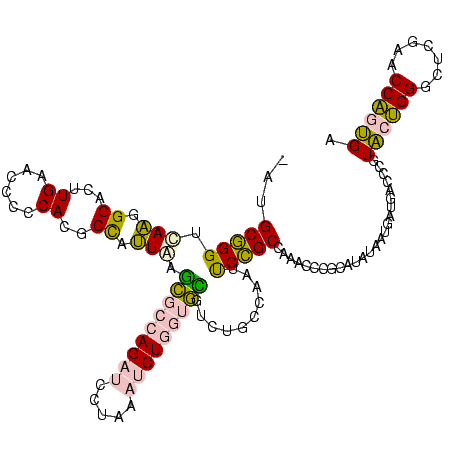
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:58:49 2006