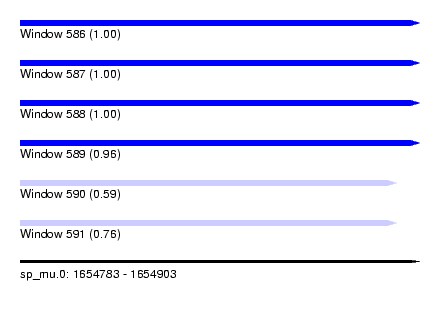
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 1,654,783 – 1,654,903 |
| Length | 120 |
| Max. P | 0.997708 |

| Location | 1,654,783 – 1,654,903 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -24.07 |
| Energy contribution | -24.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1654783 120 + 2030921/0-120 AUCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUACUCUACCAAGCUGAGCUACUUCCCGUAAAACCUCUCCUCAUCAUUAAAGUGUGCUAGUCCUUUACUAACC ..(((((((.((((.......))))......((((((.....))))))..(((.........)))...)))))))...................(((((..(....)..)))))...... ( -23.50) >sy_ha.0 2500550 107 + 2685015/0-120 GUCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAGCUGAGCUACUUCCCGUAUGUAAU-------UAAAUAAGAAAUAAAA------UUUAAAUG ..(((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))........-------................------........ ( -26.10) >sy_sa.0 2311725 104 + 2516575/0-120 GUCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAGCUGAGCUACUUCCCGUAUAUA---------UAAUUAACAUGCAUU-------UUAAAAUG ..(((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))......---------...............-------........ ( -26.10) >consensus GUCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAGCUGAGCUACUUCCCGUAUAUA_U_______UAAUUAAAAUGUAAUA______UUAAAAUG ..(((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))............................................. (-24.07 = -24.40 + 0.33)

| Location | 1,654,783 – 1,654,903 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.52 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -31.55 |
| Energy contribution | -31.33 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
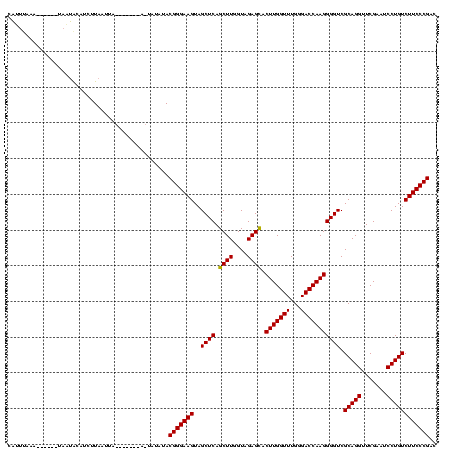
>sp_mu.0 1654783 120 + 2030921/0-120 GGUUAGUAAAGGACUAGCACACUUUAAUGAUGAGGAGAGGUUUUACGGGAAGUAGCUCAGCUUGGUAGAGUACUUGGUUUGGGACCAAGGGGUCGCAGGUUCGAAUCCUGUCUUCCCGAU .((((((.....))))))...(((((....)))))..........(((((((..((((.((((....)))).(((((((...))))))))))).(((((.......)))))))))))).. ( -36.50) >sy_ha.0 2500550 107 + 2685015/0-120 CAUUUAAA------UUUUAUUUCUUAUUUA-------AUUACAUACGGGAAGUAGCUCAGCUUGGUAGAGCACUUGGUUUGGGACCAAGGGGUCGCAGGUUCGAAUCCUGUCUUCCCGAC ........------................-------........(((((((..((((.((((....)))).(((((((...))))))))))).(((((.......)))))))))))).. ( -32.10) >sy_sa.0 2311725 104 + 2516575/0-120 CAUUUUAA-------AAUGCAUGUUAAUUA---------UAUAUACGGGAAGUAGCUCAGCUUGGUAGAGCACUUGGUUUGGGACCAAGGGGUCGCAGGUUCGAAUCCUGUCUUCCCGAC ........-------...............---------......(((((((..((((.((((....)))).(((((((...))))))))))).(((((.......)))))))))))).. ( -32.10) >consensus CAUUUAAA______UAAUACAUCUUAAUUA_______A_UAUAUACGGGAAGUAGCUCAGCUUGGUAGAGCACUUGGUUUGGGACCAAGGGGUCGCAGGUUCGAAUCCUGUCUUCCCGAC .............................................(((((((..((((.((((....)))).(((((((...))))))))))).(((((.......)))))))))))).. (-31.55 = -31.33 + -0.22) # Strand winner: reverse (1.00)

| Location | 1,654,783 – 1,654,903 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.22 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -22.22 |
| Energy contribution | -20.67 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
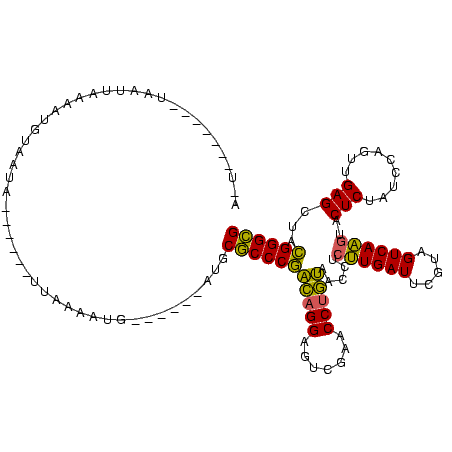
>sp_mu.0 1654783 120 + 2030921/80-200 CCUCUCCUCAUCAUUAAAGUGUGCUAGUCCUUUACUAACCCCACUCAUGCACCCUGCAGGAUUCGAACCUGCAACCGCCUGAUUCGUAGUCAGGUACUCUAUCCAGUUGAGCCAAGGGUG .................((((.(.((((.....)))).)..))))....(((((((((((.......)))))....(((((((.....))))))).(((.........)))...)))))) ( -29.80) >sy_ha.0 2500550 101 + 2685015/80-200 AAU-------UAAAUAAGAAAUAAAA------UUUAAAUG------ACGCGCCCGAUAGGAGUCGAACCCAUAACCUCUUGAUCCGUAGUCAAACGCUCUAUCCAGUUGAGCUACGGGCG ...-------................------........------...(((((....(((.((((............)))))))(((((..(((..........)))..)))))))))) ( -21.40) >sy_sa.0 2311725 98 + 2516575/80-200 A---------UAAUUAACAUGCAUU-------UUAAAAUG------GUGCGCCCGACAGGGGUCGAACCUGUAACCUCUUGAUUCGUAGUCAAGUACUCUAUCCAGUUGAGCUACGGGCG .---------..........(((((-------.......)------))))((((((((((.......))))).....((((((.....))))))..(((.........)))...))))). ( -27.40) >consensus A_U_______UAAUUAAAAUGUAAUA______UUAAAAUG______AUGCGCCCGACAGGAGUCGAACCUGUAACCUCUUGAUUCGUAGUCAAGUACUCUAUCCAGUUGAGCUACGGGCG .................................................(((((((((((.......))))).....((((((.....))))))..(((.........)))...)))))) (-22.22 = -20.67 + -1.55)

| Location | 1,654,783 – 1,654,903 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.22 |
| Mean single sequence MFE | -27.93 |
| Consensus MFE | -24.54 |
| Energy contribution | -21.67 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.40 |
| Mean z-score | -0.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
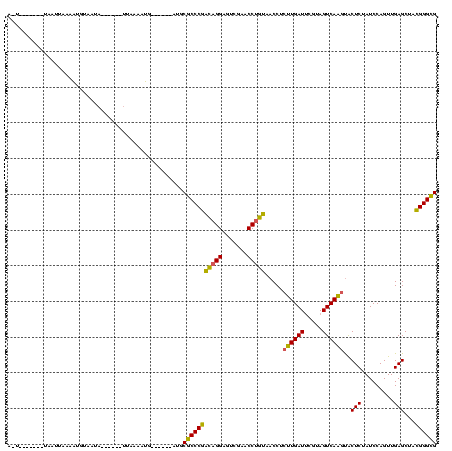
>sp_mu.0 1654783 120 + 2030921/80-200 CACCCUUGGCUCAACUGGAUAGAGUACCUGACUACGAAUCAGGCGGUUGCAGGUUCGAAUCCUGCAGGGUGCAUGAGUGGGGUUAGUAAAGGACUAGCACACUUUAAUGAUGAGGAGAGG ...((((..((((..........((((((((((.(......)..))))(((((.......))))).))))))..(((((..((((((.....)))))).)))))......))))..)))) ( -38.20) >sy_ha.0 2500550 101 + 2685015/80-200 CGCCCGUAGCUCAACUGGAUAGAGCGUUUGACUACGGAUCAAGAGGUUAUGGGUUCGACUCCUAUCGGGCGCGU------CAUUUAAA------UUUUAUUUCUUAUUUA-------AUU ((((((..((((.........))))(((.((((...((((....))))...)))).)))......))))))...------........------................-------... ( -24.00) >sy_sa.0 2311725 98 + 2516575/80-200 CGCCCGUAGCUCAACUGGAUAGAGUACUUGACUACGAAUCAAGAGGUUACAGGUUCGACCCCUGUCGGGCGCAC------CAUUUUAA-------AAUGCAUGUUAAUUA---------U .(((((..((((.........)))).(((((.......))))).....(((((.......))))))))))(((.------........-------..)))..........---------. ( -21.60) >consensus CGCCCGUAGCUCAACUGGAUAGAGUACUUGACUACGAAUCAAGAGGUUACAGGUUCGACUCCUGUCGGGCGCAU______CAUUUAAA______UAAUACAUCUUAAUUA_______A_U ((((((..((((.........)))).(((((.......))))).....(((((.......)))))))))))................................................. (-24.54 = -21.67 + -2.88) # Strand winner: reverse (0.75)

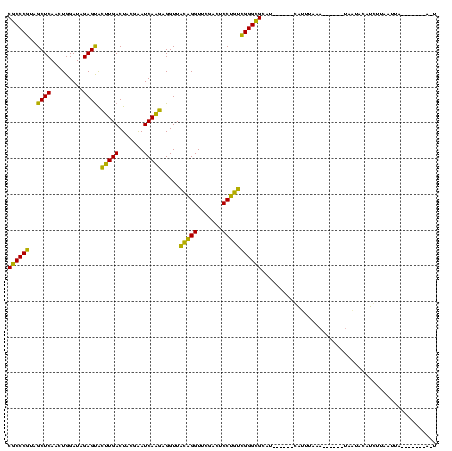
| Location | 1,654,783 – 1,654,896 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.84 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.592833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
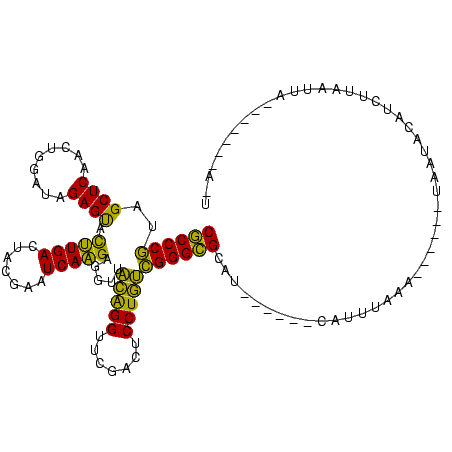
>sp_mu.0 1654783 113 + 2030921/200-320 CUCCUUCUUA---UUUAUGCCGAGGACCGGAAUCGAACCGGUACGAAGGUUCCCCUUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCCUCC---UA-AAGCGAA ..........---....(((.((((.((((....((((.((((.(((((....)))))(((((((.....))))))).....)))).)))).......)))))))).---..-..))).. ( -36.50) >sp_pn.0 1789053 109 + 2038615/200-320 CUCCAUAUUA---U----GCCGAGGACCGGAAUCGAACCGGUACGAUCGUUACCAAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCCUCU---CU-AAGCGAA ..........---.----...((((.((((....((((.((((.((.(((........(((((((.....)))))))))))))))).)))).......)))))))).---..-....... ( -32.80) >sp_ag.0 2138176 111 - 2160267/200-320 CUAAAUAUUA---U--AUGCCGAGGACCGGAAUCGAACCGGUACGAUGUUUACCA-UCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCCUCU---AACAAGCGAA ..........---.--.....((((.((((....((((.((((.(((((......-..(((((((.....)))))))))))))))).)))).......)))))))).---.......... ( -34.80) >sy_ha.0 2500550 117 + 2685015/200-320 CUUUAUUUAA--GAUGGUGCCGAGGACCGGAAUCGAACCGGUACGGUGAUCUCUCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCAAUA-UAAUGGAGCGGA (((((((...--.(((.(((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..))))))).))-)))))))).... ( -39.60) >sy_sa.0 2311725 120 + 2516575/200-320 CUAUAUUAAAAUGAUGGUGCCGAGGACCGGAAUCGAACCGGUACGAUAAUCACUUAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCAAAAAUAAUGGAGCAGA .................(((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..)))))))................ ( -32.70) >consensus CUACAUAUUA___UU_AUGCCGAGGACCGGAAUCGAACCGGUACGAUGAUUACCAAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCCUCA___AU_AAGCGAA ..................((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..))))))................. (-32.08 = -31.84 + -0.24)


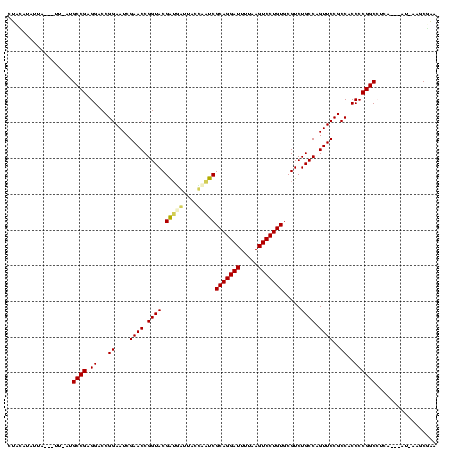
| Location | 1,654,783 – 1,654,896 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -36.34 |
| Energy contribution | -35.66 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1654783 113 + 2030921/200-320 UUCGCUU-UA---GGAGGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAAGGGGAACCUUCGUACCGGUUCGAUUCCGGUCCUCGGCAUAAA---UAAGAAGGAG ...(((.-..---.(((((((((((....(((((((........(((((.......)))))((((((....))))))..))))))).))))))).))))))).....---.......... ( -43.00) >sp_pn.0 1789053 109 + 2038615/200-320 UUCGCUU-AG---AGAGGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUUGGUAACGAUCGUACCGGUUCGAUUCCGGUCCUCGGC----A---UAAUAUGGAG ...(((.-.(---((.(((((((((....(((((((........(((((.......)))))((((((....))))))..))))))).)))))))))))))))----.---.......... ( -40.40) >sp_ag.0 2138176 111 - 2160267/200-320 UUCGCUUGUU---AGAGGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGA-UGGUAAACAUCGUACCGGUUCGAUUCCGGUCCUCGGCAU--A---UAAUAUUUAG ......((((---(...(((((((.(.(((((((((........(((((.......)))))(((-((.....)))))..))))).....)))).))))))))..--.---)))))..... ( -38.10) >sy_ha.0 2500550 117 + 2685015/200-320 UCCGCUCCAUUA-UAUUGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGGUGAGAGAUCACCGUACCGGUUCGAUUCCGGUCCUCGGCACCAUC--UUAAAUAAAG ............-...((((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).))))))))).....--.......... ( -39.10) >sy_sa.0 2311725 120 + 2516575/200-320 UCUGCUCCAUUAUUUUUGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUAAGUGAUUAUCGUACCGGUUCGAUUCCGGUCCUCGGCACCAUCAUUUUAAUAUAG ................((((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).)))))))))................. ( -36.30) >consensus UUCGCUU_AU___AGAGGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUGGGUAACCAUCGUACCGGUUCGAUUCCGGUCCUCGGCAC_AA___UAAAAUAGAG .................(((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).)))))))).................. (-36.34 = -35.66 + -0.68) # Strand winner: reverse (1.00)

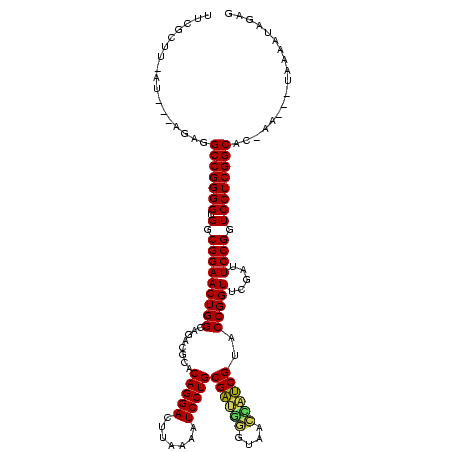
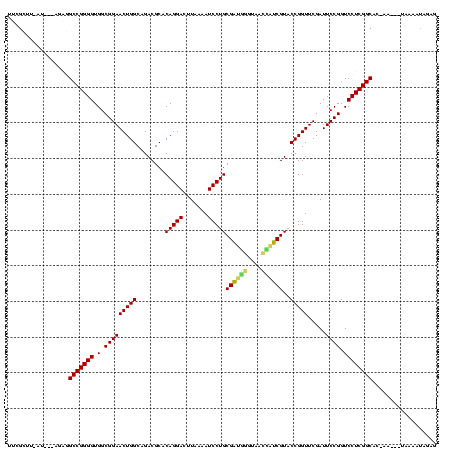
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:58:44 2006