
| Sequence ID | sy_au.0 |
|---|---|
| Location | 2,228,674 – 2,228,794 |
| Length | 120 |
| Max. P | 0.999961 |

| Location | 2,228,674 – 2,228,787 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -16.75 |
| Energy contribution | -14.87 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
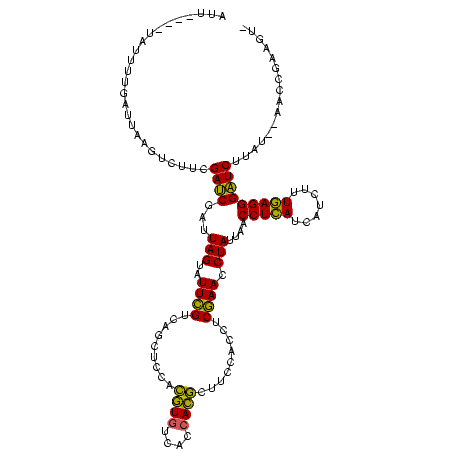
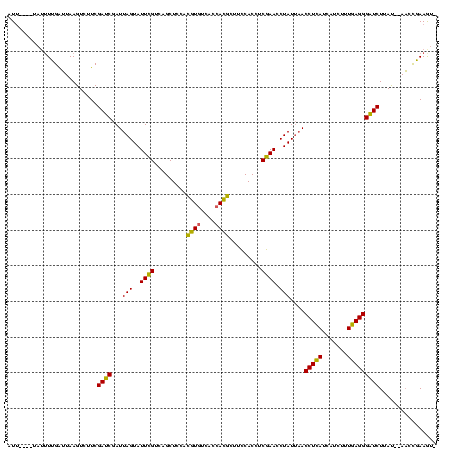
>sy_au.0 2228674 113 + 2809422/0-120 AUU----UAUUUUGAUUAAGUCUUCGAUCGAUUAGUAUUCGUCAGCUCCACAUGUCACCAUGCUUCCACCUCGAACCUAUUAACCUCAUCAUCUUUGAGGGAUCUUAU--AACCGAAGU- ...----..............(((((.....((((((((((.........((((....)))).........))))..))))))(((((.......)))))........--...))))).- ( -18.17) >sy_ha.0 1800356 117 - 2685015/0-120 AUUGAAAUAAAUUGAUUAAGUCUUCGAUCGAUUAGUAUUCGUCAGCUCCACGUAUCACCACGCUUCCACCUCGAACCUAUUAACCUCAUCAUCUUUGAGGGAUCUUAU--AACCGAAGU- .....................(((((.....((((((((((.........(((......))).........))))..))))))(((((.......)))))........--...))))).- ( -16.97) >sp_ag.0 1990286 117 - 2160267/0-120 AUC---UCUUUUUGGAUAAGUCCUCGAGCUAUUAGUAUUAGUCCGCUAAAUGUGUCACCACAAUUACACUCCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUGAUAUAAAAUC ..(---((.....(((....)))..))).((((((((..((((((.(((.((((....)))).))))...........(((.....))).........)))))..))))))))....... ( -20.90) >consensus AUU____UAUUUUGAUUAAGUCUUCGAUCGAUUAGUAUUCGUCAGCUCCACGUGUCACCACGCUUCCACCUCGAACCUAUUAACCUCAUCAUCUUUGAGGGAUCUUAU__AACCGAAGU_ .........................((((...(((..((((.........((((....)))).........)))).)))....(((((.......)))))))))................ (-16.75 = -14.87 + -1.88)

| Location | 2,228,674 – 2,228,787 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.42 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -25.17 |
| Energy contribution | -23.07 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997147 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
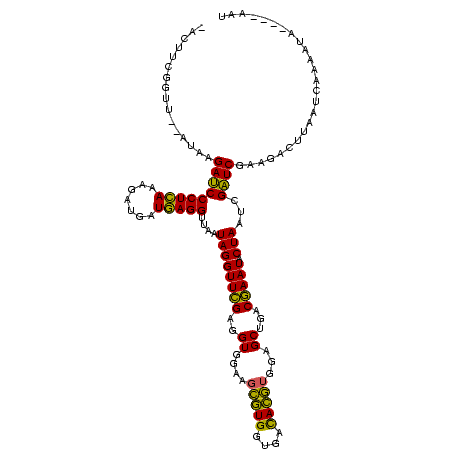
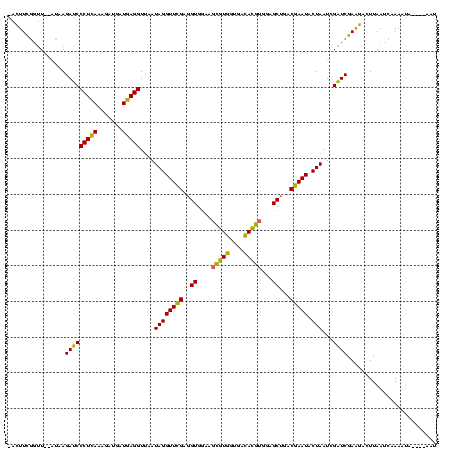
>sy_au.0 2228674 113 + 2809422/0-120 -ACUUCGGUU--AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAAUA----AAU -.((((((((--........(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))..............----... ( -32.00) >sy_ha.0 1800356 117 - 2685015/0-120 -ACUUCGGUU--AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCGUGGUGAUACGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAUUUAUUUCAAU -.((((((((--........(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))..................... ( -27.80) >sp_ag.0 1990286 117 - 2160267/0-120 GAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGGAGUGUAAUUGUGGUGACACAUUUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAAAGA---GAU ........(((.....(((((((((.......)))))....((((((((..((.(((.((((....)))).)))))...))))).)))...))))..(((....))).......---))) ( -28.60) >consensus _ACUUCGGUU__AUAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCGUGGUGACACGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAAUA____AAU ................(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))......................... (-25.17 = -23.07 + -2.10) # Strand winner: reverse (1.00)

| Location | 2,228,674 – 2,228,793 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -39.32 |
| Energy contribution | -37.33 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 119 + 2809422/120-240 UGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUGUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCA- .(..(((((...((((...(((((((((((....))))))..)))))...)))).)))))..)((((...(((((...........))))).........(((.....)))...)))).- ( -38.50) >sy_ha.0 1800356 119 - 2685015/120-240 UGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUAUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCA- .(..(((((...((((...(((((((((((....))))))..)))))...)))).)))))..)((((...(((((...........))))).........(((.....)))...)))).- ( -38.50) >sp_ag.0 1990286 120 - 2160267/120-240 UGGACUUACCGCUGGUGUACCAGUUGUCUUGCCAAAGGCAUCGCUGGGUAGCUAUGUAGGGAAGGGAUAAGCGCUGAAAGCAUCUAAGUGCGAAGCCCACCUCAAGAUGAGAUUUCCCAU .((.....))((((((...(((((((((((....))))))..)))))...)))).)).((((((((....(((((...........)))))....)))..(((.....)))..))))).. ( -40.40) >consensus UGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUAUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCA_ .((((((((...((((...(((((((((((....))))))..)))))...)))).)))))...(((....(((((...........)))))....)))..(((.....)))...)))... (-39.32 = -37.33 + -1.99) # Strand winner: reverse (0.99)

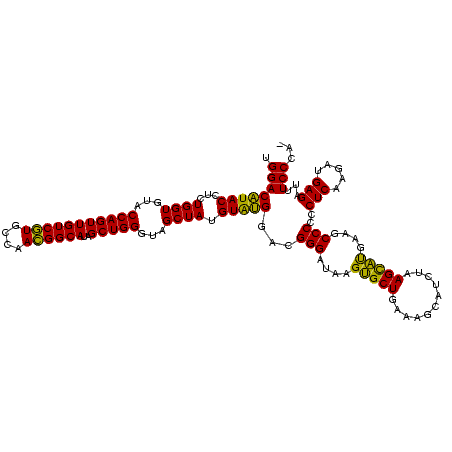
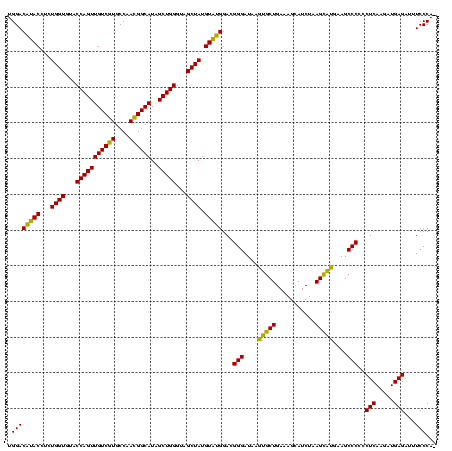
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -53.80 |
| Consensus MFE | -54.46 |
| Energy contribution | -53.80 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/280-400 CAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGG .(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).)))))........ ( -52.30) >sy_ha.0 1800356 120 - 2685015/280-400 CAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGG .(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).)))))........ ( -52.30) >sp_ag.0 1990286 120 - 2160267/280-400 CGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGG .(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).)))))........ ( -56.80) >consensus CAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGG .(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).)))))........ (-54.46 = -53.80 + -0.66) # Strand winner: reverse (1.00)

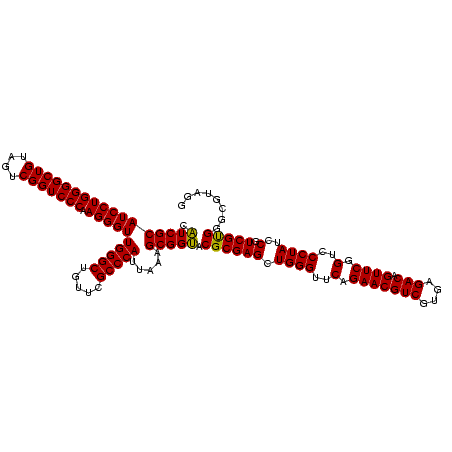
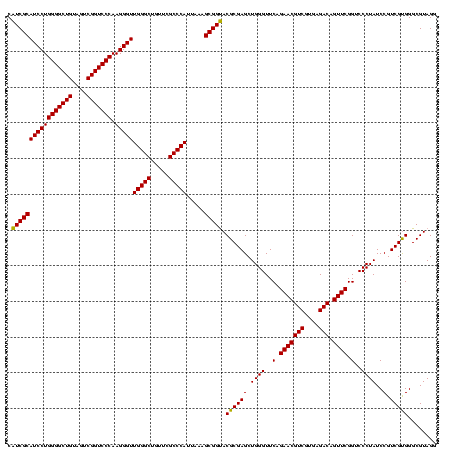
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -42.64 |
| Energy contribution | -42.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.31 |
| Structure conservation index | 1.01 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/320-440 GUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAAC ((((........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))....)))) ( -42.00) >sy_ha.0 1800356 120 - 2685015/320-440 GUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAAC ((((........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))....)))) ( -42.00) >sp_ag.0 1990286 120 - 2160267/320-440 GUUCUGAACCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAAC ((((........((((((((..........(((((.....)))))...((((((.(........).))))))..))).)))))((((...(((((.....)))))...))))....)))) ( -43.00) >consensus GUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAAC ((((........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))....)))) (-42.64 = -42.20 + -0.44)

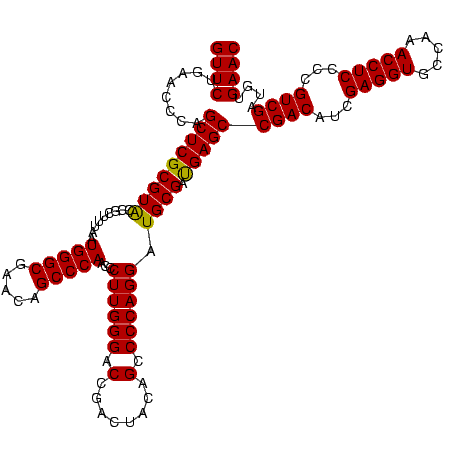
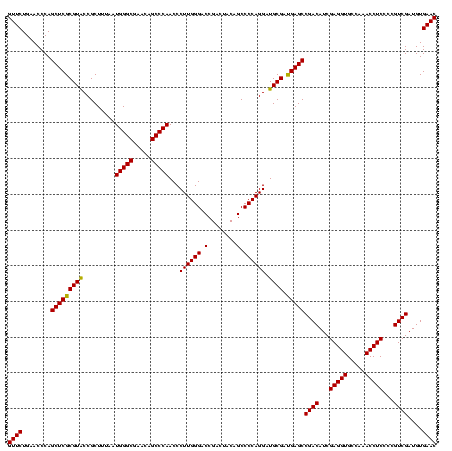
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -52.97 |
| Consensus MFE | -53.28 |
| Energy contribution | -52.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.81 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/320-440 GUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAAC ((((....(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))........)))) ( -52.30) >sy_ha.0 1800356 120 - 2685015/320-440 GUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAAC ((((....(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))........)))) ( -52.30) >sp_ag.0 1990286 120 - 2160267/320-440 GUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAAC ((((....(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....))))))........)))) ( -54.30) >consensus GUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAAC ((((....(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))........)))) (-53.28 = -52.83 + -0.44) # Strand winner: reverse (1.00)

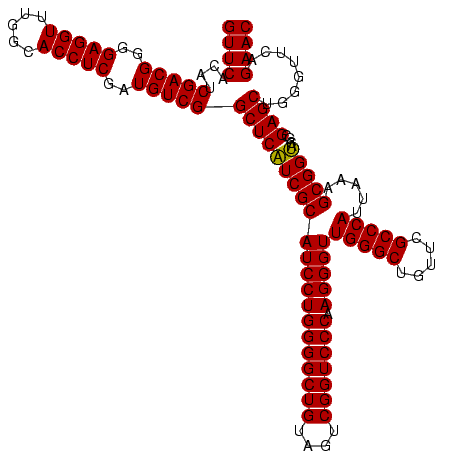
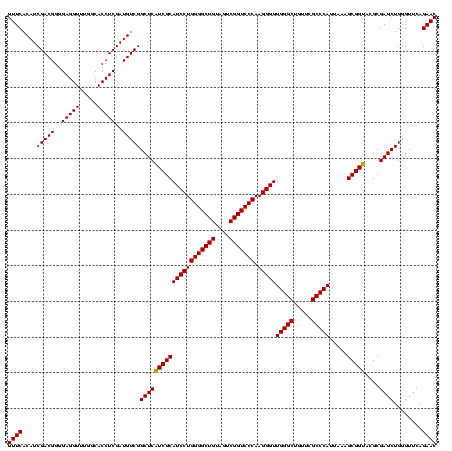
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -48.47 |
| Consensus MFE | -47.80 |
| Energy contribution | -47.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/360-480 AAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGC ............((((((((.....))))))))(((((((((..(((((((.((............)))))))))..))))......(((((((((((....))))))).))))))))). ( -47.90) >sy_ha.0 1800356 120 - 2685015/360-480 AAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGC ............((((((((.....))))))))(((((((((..(((((((.((............)))))))))..))))......(((((((((((....))))))).))))))))). ( -47.90) >sp_ag.0 1990286 120 - 2160267/360-480 AAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGC ....((((((((.(((((.(((((((.((((((((...((.......))...))))))))..))).((((((((.(((....)))))))..)))))))).....))))).)))))..))) ( -49.60) >consensus AAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGC ............((((((((.....))))))))(((((((((..(((((((.((............)))))))))..))))......(((((((((((....))))))).))))))))). (-47.80 = -47.80 + 0.00) # Strand winner: reverse (0.97)


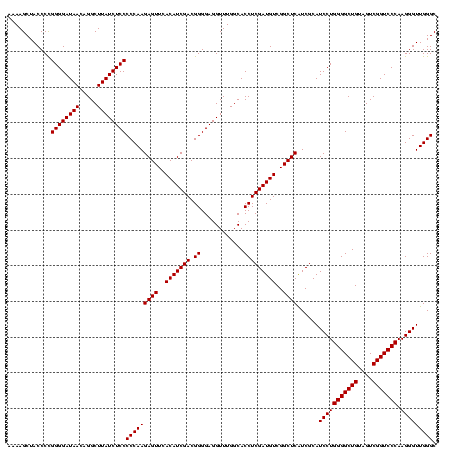
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -39.22 |
| Energy contribution | -37.67 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/520-640 UAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUG ..((.(((..((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))((((............))))))))...(((....)))...))).)). ( -38.70) >sy_ha.0 1800356 120 - 2685015/520-640 UAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUG ..((.(((..((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))((((............))))))))...(((....)))...))).)). ( -38.70) >sp_ag.0 1990286 120 - 2160267/520-640 UAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUG ..........((((..(((((((((..(((((((.....)))))))......((((....)))))))))))))..((((..(((......)))..)))).(((....)))))))...... ( -44.60) >consensus UAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUG ..((.(((.(......(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))).).))).)). (-39.22 = -37.67 + -1.55) # Strand winner: reverse (1.00)

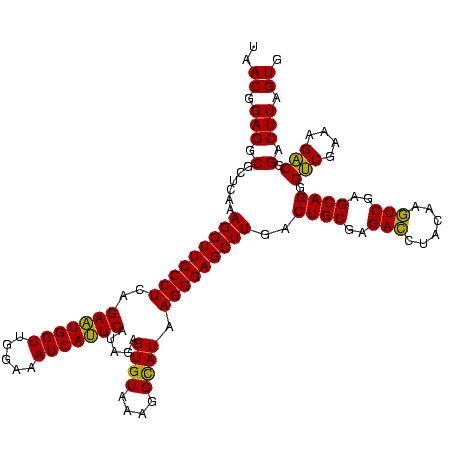
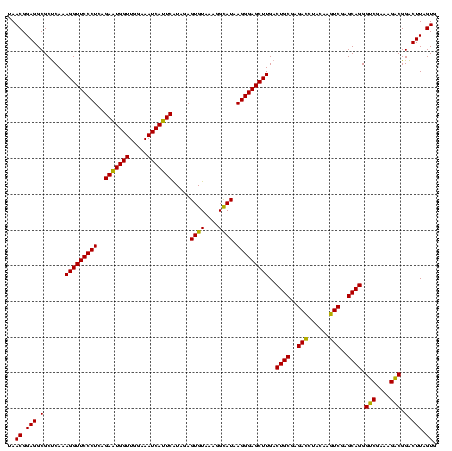
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -33.66 |
| Consensus MFE | -33.10 |
| Energy contribution | -32.44 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/560-680 CGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUG .(((((...((((.......((((((........(((((.........))))))))))).......))))((((((...........))))))(((.......)))..)))))....... ( -32.34) >sy_ha.0 1800356 120 - 2685015/560-680 CGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUG .(((((...((((.......((((((........(((((.........))))))))))).......))))((((((...........))))))(((.......)))..)))))....... ( -32.34) >sp_ag.0 1990286 120 - 2160267/560-680 CGCAGUCAAGCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAG .(((((...(((((((((................(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))....... ( -36.29) >consensus CGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUG .(((((...((((.......((((((........(((((.........))))))))))).......))))((((((...........))))))(((.......)))..)))))....... (-33.10 = -32.44 + -0.66)

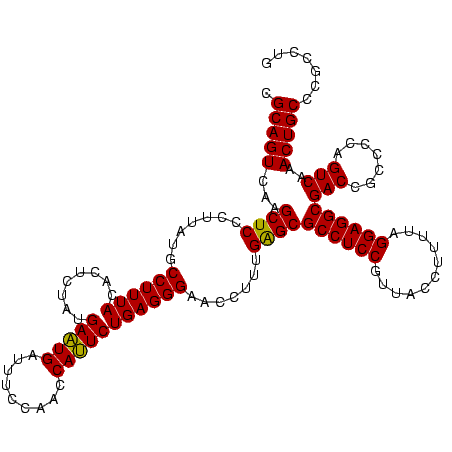
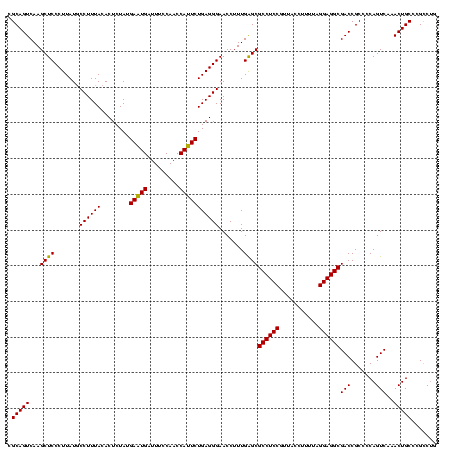
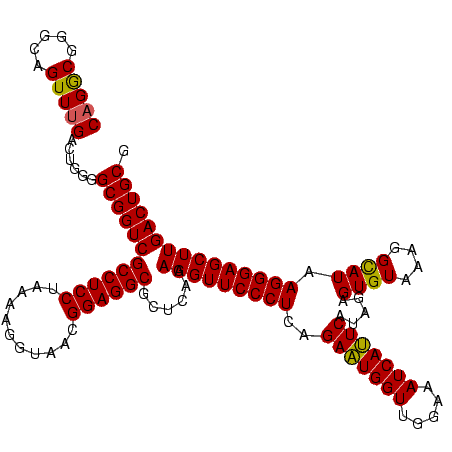
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -45.23 |
| Consensus MFE | -44.15 |
| Energy contribution | -43.60 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/560-680 CAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCG (((((.....)))))......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).))))))))))))))). ( -45.20) >sy_ha.0 1800356 120 - 2685015/560-680 CAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCG (((((.....)))))......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).))))))))))))))). ( -45.20) >sp_ag.0 1990286 120 - 2160267/560-680 CUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCG .......((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....)))))))......((((....))))))))))))))))))). ( -45.30) >consensus CAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCG (((((.....)))))......((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).))))))))))))))). (-44.15 = -43.60 + -0.55) # Strand winner: reverse (1.00)

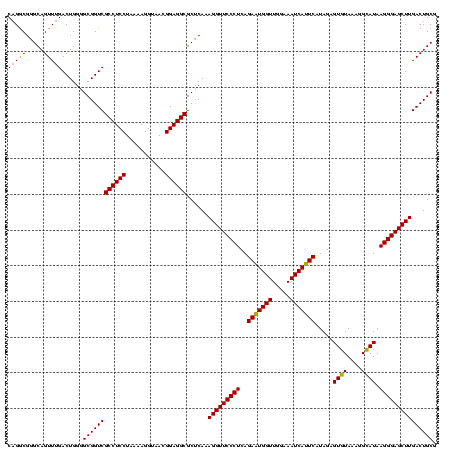
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -39.01 |
| Energy contribution | -36.57 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/600-720 UUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAG ............(((.(((......((..(((((((...........)))))))..)))))))).....((((((((..(((...)))....(((((.....)))))))))).))).... ( -39.20) >sy_ha.0 1800356 120 - 2685015/600-720 UUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAG ............(((.(((......((..(((((((...........)))))))..)))))))).....((((((((..(((...)))....(((((.....)))))))))).))).... ( -39.20) >sp_ag.0 1990286 120 - 2160267/600-720 UUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUA .(((.((((.(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).........((((((.....))))))..)))).))).. ( -46.40) >consensus UUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAG ............(((.(((..........(((((((...........)))))))....)))))).....(((((((...(((...)))...((((((.....)))))))))).))).... (-39.01 = -36.57 + -2.44)

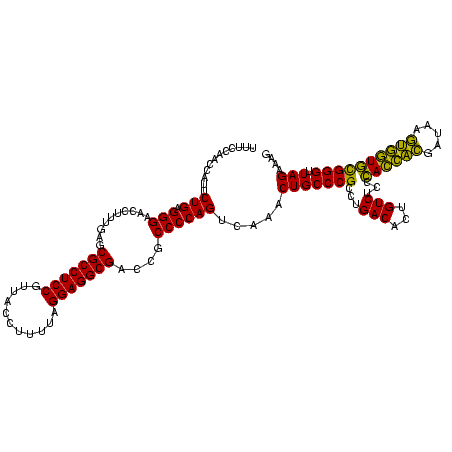
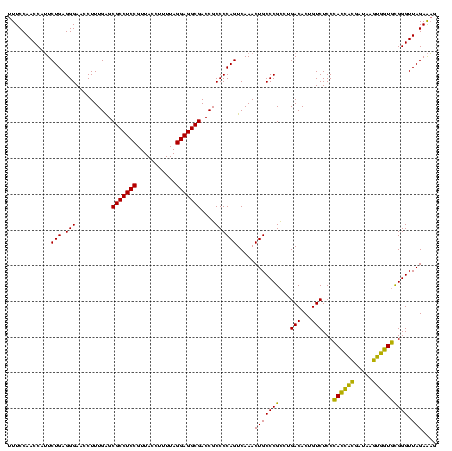
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -48.10 |
| Consensus MFE | -47.86 |
| Energy contribution | -45.53 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/600-720 CUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAA ..((((((((..((((((.....)))))).(((.....(((((((.....))))))).(((((((.(((((((...........))))))))).....)))))))).....)))))))). ( -47.30) >sy_ha.0 1800356 120 - 2685015/600-720 CUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAA ..((((((((..((((((.....)))))).(((.....(((((((.....))))))).(((((((.(((((((...........))))))))).....)))))))).....)))))))). ( -47.30) >sp_ag.0 1990286 120 - 2160267/600-720 UACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAA ...(((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).))))))).. ( -49.70) >consensus CUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAA ...(((((((..((((((.....)))))).(((.....(((((((.....))))))).(((((((.(((((((...........))))))))).....)))))))).....))))))).. (-47.86 = -45.53 + -2.32) # Strand winner: reverse (1.00)

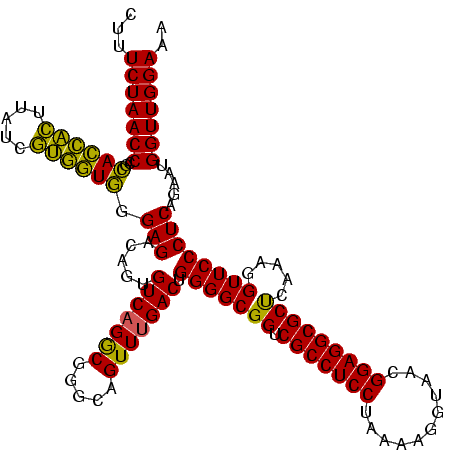
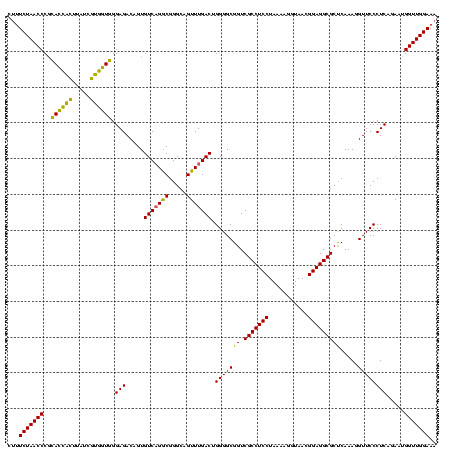
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -41.43 |
| Consensus MFE | -40.79 |
| Energy contribution | -39.13 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/640-760 CCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGU .......(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))).... ( -44.30) >sy_ha.0 1800356 120 - 2685015/640-760 CCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGU .......(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))).... ( -44.30) >sp_ag.0 1990286 120 - 2160267/640-760 CCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACG ........((((((((.......)))..(((((((........(((.(((.((((((.....)))))).))).))).((.......))....)))))))...........)))))..... ( -35.70) >consensus CCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGU .......(((((((((.......)))..(((((((........(((.(((.((((((.....)))))).))).)))..(((.......))).)))))))...........)))))).... (-40.79 = -39.13 + -1.66)

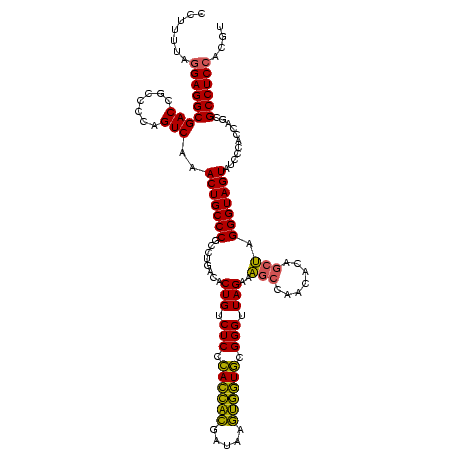
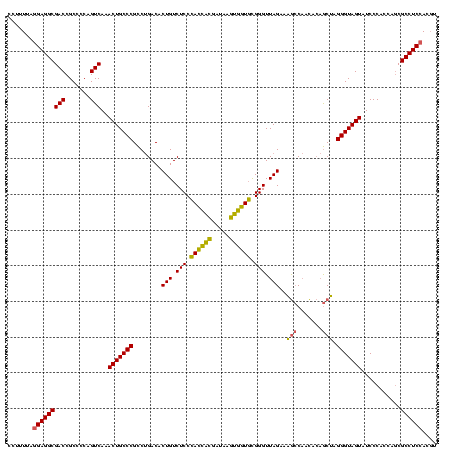
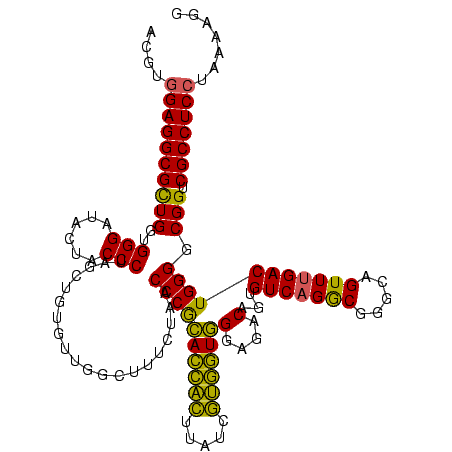
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.78 |
| Mean single sequence MFE | -46.83 |
| Consensus MFE | -43.81 |
| Energy contribution | -41.60 |
| Covariance contribution | -2.21 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/640-760 ACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGG ....((((((((((((((....))))((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))).))).)))))))....... ( -52.10) >sy_ha.0 1800356 120 - 2685015/640-760 ACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGG ....((((((((((((((....))))((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))).))).)))))))....... ( -52.10) >sp_ag.0 1990286 120 - 2160267/640-760 CGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGG .....((((((((((.(((......)))....................(((((.(((((....((..(((((.......)))))..))..))))).))))))))).))))))........ ( -36.30) >consensus ACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGG ....((((((((((..(((......)))....................((((((((((.....))))))(....)...(((((((.....))))))))))).))).)))))))....... (-43.81 = -41.60 + -2.21) # Strand winner: reverse (0.99)

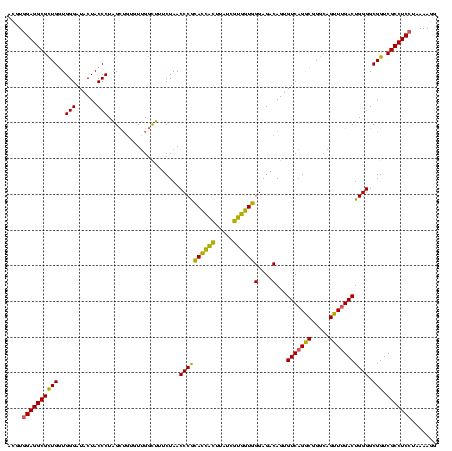
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -34.53 |
| Consensus MFE | -33.41 |
| Energy contribution | -29.53 |
| Covariance contribution | -3.88 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/680-800 ACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGU ...(((.(((.((((((.....)))))).))).))).........((((...((((((..........(.((((..((((.(((....))).)))).....)))).)))))))...)))) ( -34.90) >sy_ha.0 1800356 120 - 2685015/680-800 ACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGU ...(((.(((.((((((.....)))))).))).))).........((((...((((((..........(.((((..((((.(((....))).)))).....)))).)))))))...)))) ( -34.90) >sp_ag.0 1990286 120 - 2160267/680-800 ACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGU ...........((((((....((((((...(((((...)))))(((((....(((......)))....(((((............)))))...))))).)))))).....)))))).... ( -33.80) >consensus ACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGU ...........((((((.....))))))((((.(((................(((......)))......((((..((((.(((....))).)))).....))))......))).)))). (-33.41 = -29.53 + -3.88)

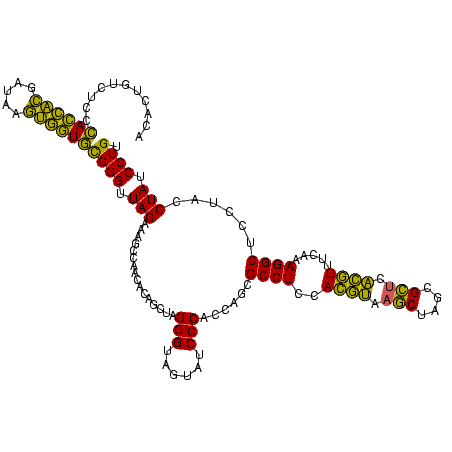
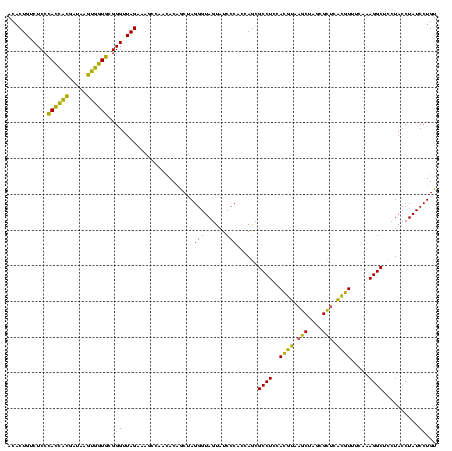
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.89 |
| Mean single sequence MFE | -46.37 |
| Consensus MFE | -42.68 |
| Energy contribution | -40.70 |
| Covariance contribution | -1.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/680-800 ACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGU .(((.((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).)))).)))(((.(((...((.((((((.....))))))))))).))).. ( -51.40) >sy_ha.0 1800356 120 - 2685015/680-800 ACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGU .(((.((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).)))).)))(((.(((...((.((((((.....))))))))))).))).. ( -51.40) >sp_ag.0 1990286 120 - 2160267/680-800 ACAGGAUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGU ...((.(((((((..((((.....((((..((....))..))))..))))......(((......)))...........)))).))).))..((((((.....))))))(....)..... ( -36.30) >consensus ACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGU .(((.((((.((((.(((((....((((((((....)))))))).)))))....((((....)))))))).)))).)))(((.(((...((.((((((.....))))))))))).))).. (-42.68 = -40.70 + -1.98) # Strand winner: reverse (1.00)

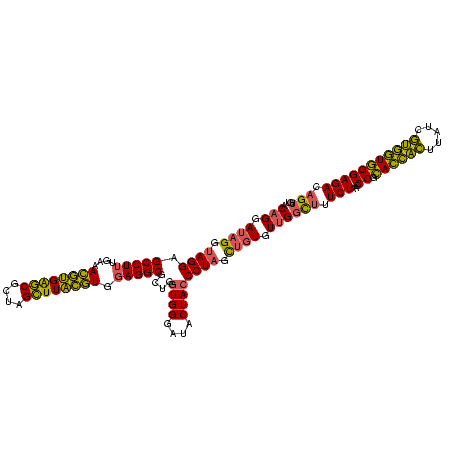
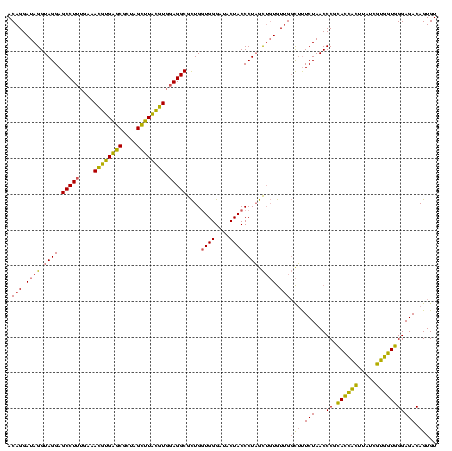
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -26.28 |
| Energy contribution | -23.40 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.929005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/720-840 CCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGGCUACAGUAAAGC .....((((...((((((..........(.((((..((((.(((....))).)))).....)))).)))))))...))))....((((((((..((......))..))).)))))..... ( -30.70) >sy_ha.0 1800356 120 - 2685015/720-840 CCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGGCUACAGUAAAGC .....((((...((((((..........(.((((..((((.(((....))).)))).....)))).)))))))...))))....((((((((..((......))..))).)))))..... ( -30.70) >sp_ag.0 1990286 120 - 2160267/720-840 GCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGUACAUGUGGUACAGAUACUCAAUAUCAAACUGCAGUAAAGC ((((((((....(((......)))....(((((............)))))...)))))....)))...........((((((.....)))))).((((((.........)).)))).... ( -25.90) >consensus CCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCUGUGCCGAAUUUCAAUAUCAGGCUACAGUAAAGC ....(((((((.(((......)))......((((..((((.(((....))).)))).....))))..................))))))).............................. (-26.28 = -23.40 + -2.88)

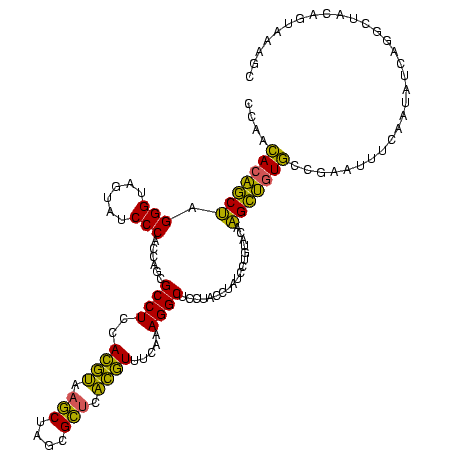

| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.67 |
| Mean single sequence MFE | -40.77 |
| Consensus MFE | -38.52 |
| Energy contribution | -34.87 |
| Covariance contribution | -3.65 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/720-840 GCUUUACUGUAGCCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGG ......((((.(((.(((......))).)))))))......(((.((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).)))).))). ( -45.30) >sy_ha.0 1800356 120 - 2685015/720-840 GCUUUACUGUAGCCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGG ......((((.(((.(((......))).)))))))......(((.((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).)))).))). ( -45.30) >sp_ag.0 1990286 120 - 2160267/720-840 GCUUUACUGCAGUUUGAUAUUGAGUAUCUGUACCACAUGUACAGGAUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGC ....((((.((((.....))))))))(((((((.....)))))))((((((....)))))).((((((.((((((............))))))...(((......)))...))))))... ( -31.70) >consensus GCUUUACUGUAGCCUGAUAUUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGG (((.......)))..............((((((((((...............((.(((((....((((((((....)))))))).))))).))(((((....)))))..)))))))))). (-38.52 = -34.87 + -3.65) # Strand winner: reverse (1.00)

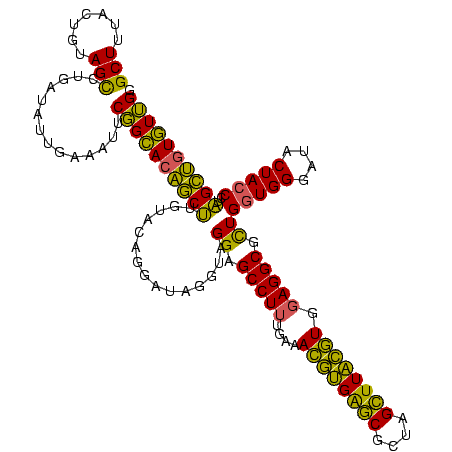
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -47.47 |
| Consensus MFE | -49.24 |
| Energy contribution | -47.47 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.38 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1000-1120 GGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGU (((((((((...(((....)))...))..))))))).(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).))) ( -48.50) >sy_ha.0 1800356 120 - 2685015/1000-1120 GGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGU (((((((((...(((....)))...))..))))))).(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).))) ( -48.50) >sp_ag.0 1990286 120 - 2160267/1000-1120 GCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAGCGUAAGCGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGU (((((((((...(((....)))...))..))))))).(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).))) ( -45.40) >consensus GGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAGCUUCUGCGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGU (((((((((...(((....)))...))..))))))).(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).))) (-49.24 = -47.47 + -1.77) # Strand winner: reverse (0.99)


| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.39 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -34.83 |
| Energy contribution | -33.67 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881773 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1080-1200 GGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACG ........(.(.(((((((.((((....)))).)))))))).)........(((((.((.((((((((.....))))...)))).))((...(((....)))...))....))))).... ( -37.90) >sy_ha.0 1800356 120 - 2685015/1080-1200 GGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACG ........(.(.(((((((.((((....)))).)))))))).)........(((((.((.((((((((.....))))...)))).))((...(((....)))...))....))))).... ( -38.60) >sp_ag.0 1990286 109 - 2160267/1080-1200 GGGAGAAGGGGCGCUGAUU-----------UUAGAUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACG ..........(((((((((-----------...)))))).))).((((((((((....))..))))))))......(((..((((.(((...(((....)))...))).)))).)))... ( -28.70) >consensus GGGAGAAGGGGUGCUCUUUAGGGU__A_GCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACG ((..(...(((((((((((.((((....)))).)))))))..(........)))))...)..))((((.....))))...(((((((((...(((....)))...))..))))))).... (-34.83 = -33.67 + -1.16) # Strand winner: reverse (1.00)

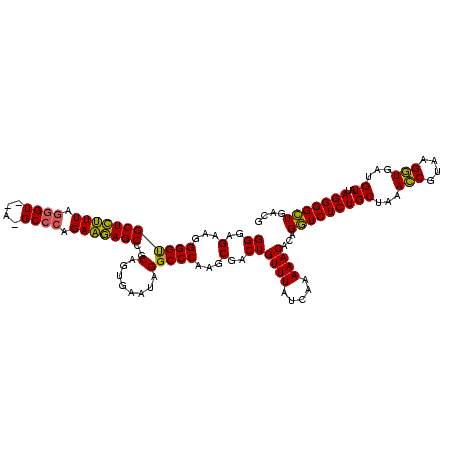
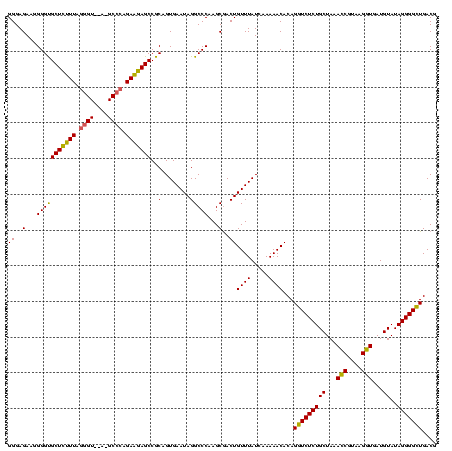
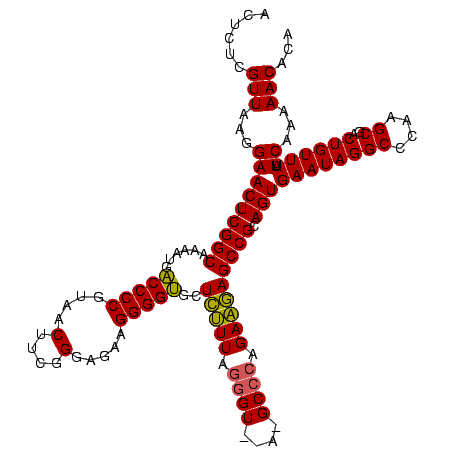
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -33.90 |
| Energy contribution | -33.40 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881065 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1120-1240 ACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACA ....(((((..((.(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).))).......))..))))).(((((.....))))).. ( -36.80) >sy_ha.0 1800356 120 - 2685015/1120-1240 ACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACA ....(((((..((.(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).))).......))..))))).(((((.....))))).. ( -37.50) >sp_ag.0 1990286 109 - 2160267/1120-1240 ACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGAUU-----------UUAGAUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACA .((((((..(((..((..(((......)))..))..))))))))).....(((((((((-----------...)))))).))).((((((((((....))..)))))))).......... ( -30.00) >consensus ACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGU__A_GCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACA ......(((...(((((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).)))((((((((....))..)))))).))...)))... (-33.90 = -33.40 + -0.50) # Strand winner: reverse (0.88)

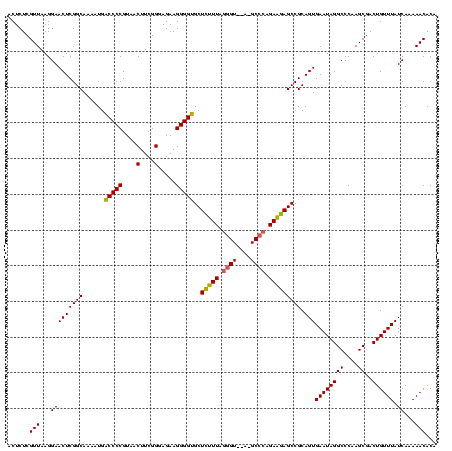
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -26.95 |
| Energy contribution | -26.37 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1160-1280 GCUCUUCUGGGCGUUAACCCUAAAGAGCACCCCUUCUCCCGAAGUUACGGGGUCAUUUUGCCGAGUUCCUUAACGAGAGUUCGCUCGCUCACCUUAGAAUUCUCAUCUUGACUACCUGUG ((((((..(((......)))..))))))....((((....)))).((((((((((...((..((((((..(((.(.((((......)))).).))))))))).))...))))..)))))) ( -34.60) >sy_ha.0 1800356 120 - 2685015/1160-1280 GCUCUUCUGGGCUUGCACCCUAAAGAGCACCCCUUCUCCCGAAGUUACGGGGUCAUUUUGCCGAGUUCCUUAACGAGAGUUCGCUCGCUCACCUUAGAAUUCUCAUCUUGACUACCUGUG ((((((..(((......)))..))))))....((((....)))).((((((((((...((..((((((..(((.(.((((......)))).).))))))))).))...))))..)))))) ( -34.60) >sp_ag.0 1990286 109 - 2160267/1160-1280 GCUGAUCUAA-----------AAUCAGCGCCCCUUCUCCCGAAGUUACGGGGCCAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGCUCACCUGAGGCUACUCGCCUCGACUACCUGUG ((((((....-----------.))))))....((((....)))).((((((((......))).((((......(((((....))))).......(((((.....)))))))))..))))) ( -32.70) >consensus GCUCUUCUGGGC_U__ACCCUAAAGAGCACCCCUUCUCCCGAAGUUACGGGGUCAUUUUGCCGAGUUCCUUAACGAGAGUUCGCUCGCUCACCUUAGAAUUCUCAUCUUGACUACCUGUG ((((((..(((......)))..))))))....((((....)))).((((((...........((((.....(((....))).....)))).....(((.......)))......)))))) (-26.95 = -26.37 + -0.58)

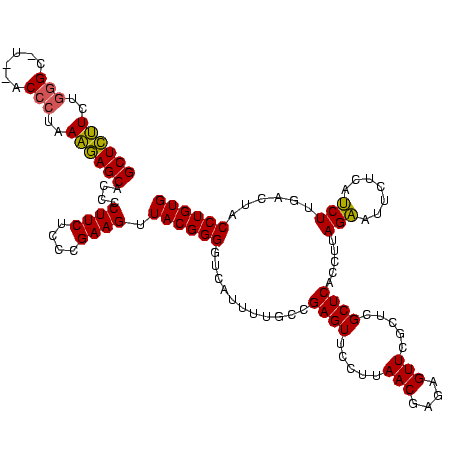
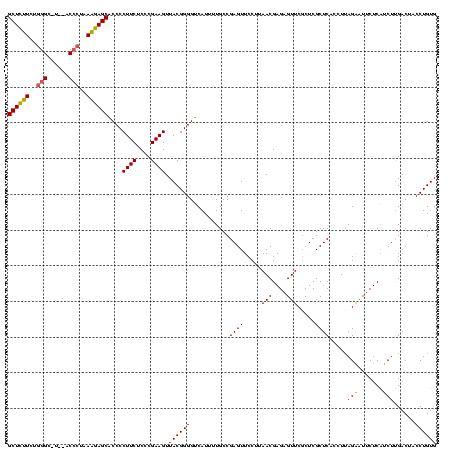
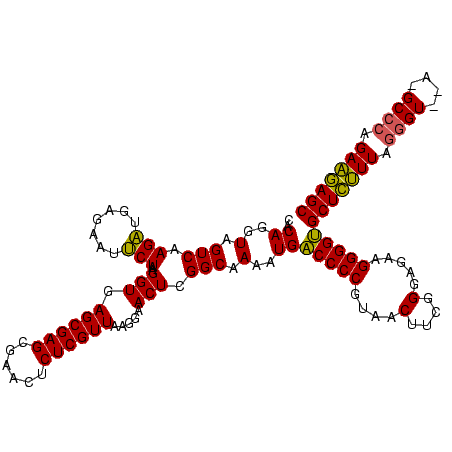
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.17 |
| Mean single sequence MFE | -38.40 |
| Consensus MFE | -35.91 |
| Energy contribution | -34.97 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1160-1280 CACAGGUAGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGC ..((..(.(((.(((.......)))..(((.((((((......)))))).....))).))).)..))(((((....(....).....)))))(((((((.((((....)))).))))))) ( -35.90) >sy_ha.0 1800356 120 - 2685015/1160-1280 CACAGGUAGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGC ..((..(.(((.(((.......)))..(((.((((((......)))))).....))).))).)..))(((((....(....).....)))))(((((((.((((....)))).))))))) ( -36.60) >sp_ag.0 1990286 109 - 2160267/1160-1280 CACAGGUAGUCGAGGCGAGUAGCCUCAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGAUU-----------UUAGAUCAGC ..((..(.((((((((.....))))).(((.(((((((....))))))).....))).))).)..))(((((....(....).....)))))(((((((-----------...))))))) ( -42.70) >consensus CACAGGUAGUCAAGAUGAGAAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGU__A_GCCCAGAAGAGC ..((..(.(((.(((.......)))..(((.((((((......)))))).....))).))).)..))(((((....(....).....)))))(((((((.((((....)))).))))))) (-35.91 = -34.97 + -0.94) # Strand winner: reverse (1.00)


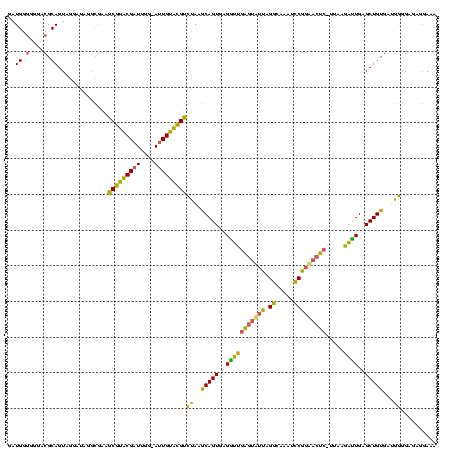
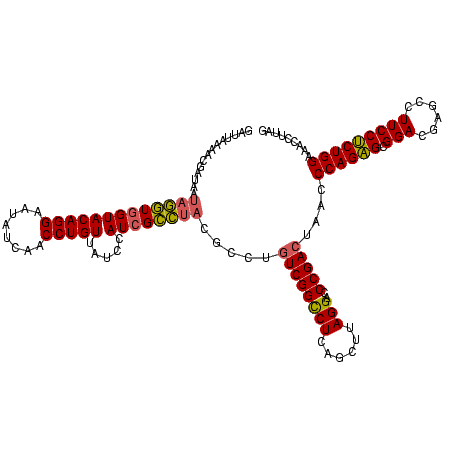
| Location | 2,228,674 – 2,228,793 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.54 |
| Mean single sequence MFE | -33.27 |
| Consensus MFE | -26.45 |
| Energy contribution | -23.47 |
| Covariance contribution | -2.98 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 119 + 2809422/1360-1480 GAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUGCGAUUGG-AUUGCACGUCUAAGCAGUAAGGCUGAGUAUUAGGCAAAUCCGGUACUCGUUAAGGCUGAGCUGUGAUGGGGAGAAGACA ..((.(....).))((.((((..((...(((((((((...-)))))))))((((.(((.....)))....))))))..))))....(((((((.(((...))).)))))))......)). ( -35.90) >sy_ha.0 1800356 118 - 2685015/1360-1480 GAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGAUUGG-AUUGUACGUCUAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAA ..((.(....).))..............(((((((((...-)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..))).......... ( -30.60) >sp_ag.0 1990286 120 - 2160267/1360-1480 GAUGGAGGGACGCAGUAGGCUAACUAAACCAGACGAUUGGAAGUGUCUGGUCAAACAGUGAGGUGUGAUAUGAGUCAAAUGCUUAUAUCUUUAACAUUGAGCUGUGACGAGGAGCGAAGU ..........(((..(((.....))).((((((((........))))))))...(((((..((((((((((((((.....)))))))))....)))))..)))))........))).... ( -33.30) >consensus GAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGAUUGG_AUUGUACGUCUAAGCAGUGAGGUUGAGUAUUAGGCAAAUCCGGAACUC_UUAAGAUUGAGCUGUGAUGGGGAGAGGAAA ..((.(....).))..............(((((((((....)))))))))((..(((((..(((((((((((.((.....))))))))).....))))..)))))...)).......... (-26.45 = -23.47 + -2.98) # Strand winner: reverse (1.00)


| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -29.18 |
| Energy contribution | -28.97 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.89 |
| Structure conservation index | 0.92 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1480-1600 GAUUAAAACGAUUAUAGGUGGUACAGGAAUAUCAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAG ..............(((((((((((((........))))).....)))))))).....((((((((......)))..)))))....((((((.(((......)))))))))......... ( -33.20) >sy_ha.0 1800356 120 - 2685015/1480-1600 GAUUAAAACGAUGCUAGGUGGUACAGGAAUAUCAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAG ............(((((((((((((((........))))).....)))))))).))..((((((((......)))..)))))....((((((.(((......)))))))))......... ( -35.40) >sp_ag.0 1990286 113 - 2160267/1480-1600 -------ACUAUAUACUCUAGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAG -------...........(((((((((........))))).......(((....((((...((.....))..)))).)))))))..((((((.(((......)))))))))......... ( -27.00) >consensus GAUUAAAACGAUAAUAGGUGGUACAGGAAUAUCAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAG ..............(((((((((((((........))))).....)))))))).....((((((((......)))..)))))....((((((.(((......)))))))))......... (-29.18 = -28.97 + -0.22)

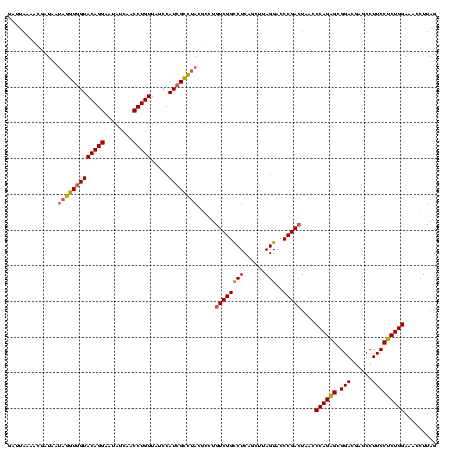
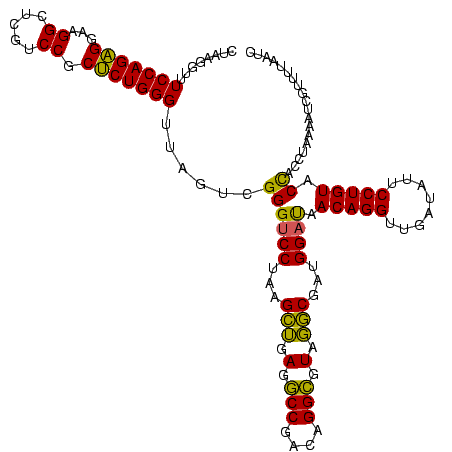
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -38.43 |
| Consensus MFE | -35.85 |
| Energy contribution | -33.97 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1480-1600 CUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUUAAUC ...((((.(((((((...((.....)).)))))))......((((((...(((.(.(((....))).).)))...)))).(((((........))))).))))))............... ( -38.00) >sy_ha.0 1800356 120 - 2685015/1480-1600 CUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAGCAUCGUUUUAAUC ....(((..((((((...((.....)).))))))(((((..((((((...(((.(.(((....))).).)))...)))).(((((........))))).))..))))))))......... ( -40.00) >sp_ag.0 1990286 113 - 2160267/1480-1600 CUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUAGAGUAUAUAGU------- ........(((((((...((.....)).)))))))((((((((((((...(((.(.(((....))).).)))...)).(((....)))....))))).)))))..........------- ( -37.30) >consensus CUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAAAAUCGUUUUAAUC ........(((((((...((.....)).)))))))......((((((...(((.(.(((....))).).)))...)))).(((((........))))).))................... (-35.85 = -33.97 + -1.88) # Strand winner: reverse (1.00)

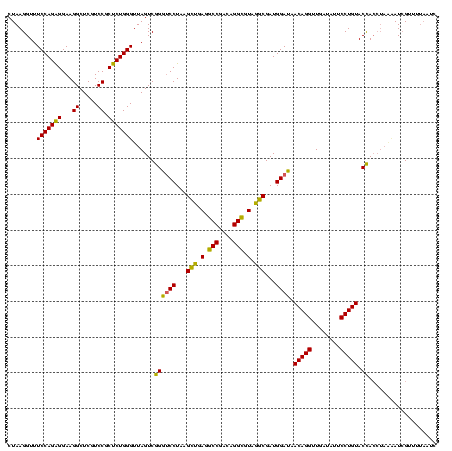
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -36.97 |
| Consensus MFE | -35.79 |
| Energy contribution | -34.47 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1520-1640 UAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUC ........((...((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))....))..... ( -36.60) >sy_ha.0 1800356 120 - 2685015/1520-1640 UAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUC ........((...((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))....))..... ( -36.60) >sp_ag.0 1990286 120 - 2160267/1520-1640 UGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUC .((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...))))))))(((((........)))))......... ( -37.70) >consensus UAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUC .............((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))........... (-35.79 = -34.47 + -1.33)

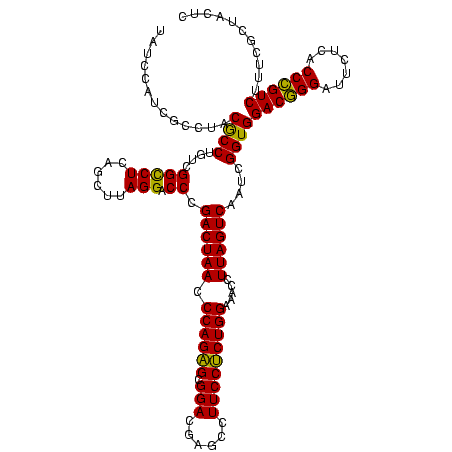
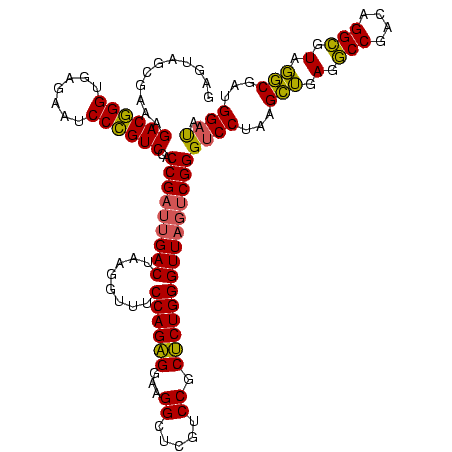
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -45.17 |
| Energy contribution | -44.07 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.14 |
| SVM RNA-class probability | 0.921164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1520-1640 GAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUA ...........((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...)))). ( -46.10) >sy_ha.0 1800356 120 - 2685015/1520-1640 GAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUA ...........((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...)))). ( -46.10) >sp_ag.0 1990286 120 - 2160267/1520-1640 GAGUAGCGCAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACA .....(((...((((((.......))))))...)))..((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))... ( -43.70) >consensus GAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUA ...........((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...)))). (-45.17 = -44.07 + -1.10) # Strand winner: reverse (1.00)


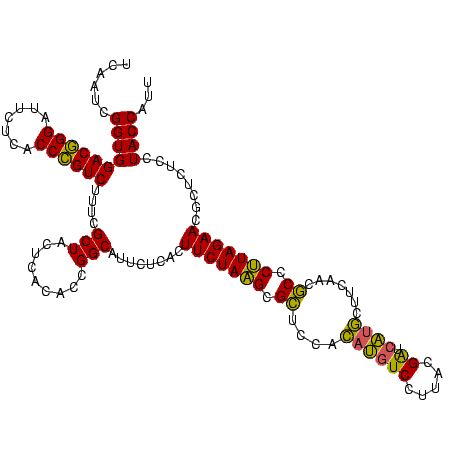
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -23.50 |
| Energy contribution | -21.73 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1600-1720 UCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUU ....((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))............... ( -25.90) >sy_ha.0 1800356 120 - 2685015/1600-1720 UCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUU ....((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))............... ( -25.90) >sp_ag.0 1990286 120 - 2160267/1600-1720 UCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUG ......((((((((((.......))))))..(((((((.((((..((........))..)))).))...)))))...........))))((((........))))............... ( -25.60) >consensus UCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUU ......((((((((((.......))))))....(((.........)))........(((((((.((....((((((.....)).)))).......)).)))))))........))))... (-23.50 = -21.73 + -1.77)

| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -42.36 |
| Energy contribution | -39.37 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.28 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998848 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1600-1720 AAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGA ...((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))((((.((....(....).((((.......))))))))))...... ( -40.80) >sy_ha.0 1800356 120 - 2685015/1600-1720 AAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGA ...((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))((((.((....(....).((((.......))))))))))...... ( -41.80) >sp_ag.0 1990286 120 - 2160267/1600-1720 CAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGCAAGACAGGUGAGAAUCCUGUCCACCGUAAGA ..(((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))).........(((...((((((.......))))))...))).... ( -42.20) >consensus AAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGA ...((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))((((............((((((.......))))))))))...... (-42.36 = -39.37 + -2.99) # Strand winner: reverse (1.00)

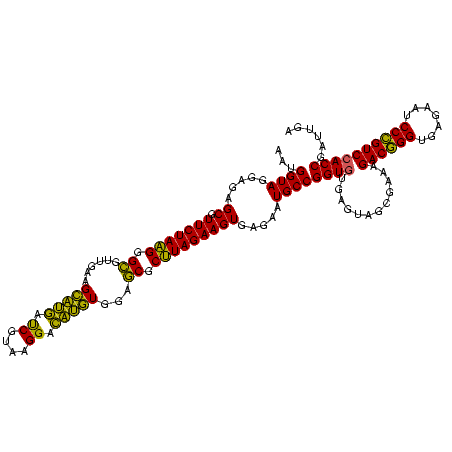
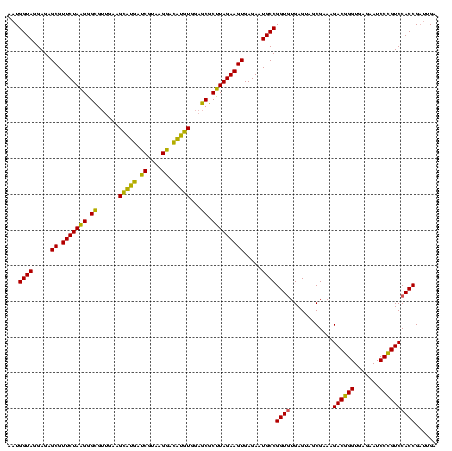
| Location | 2,228,674 – 2,228,792 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -17.76 |
| Energy contribution | -15.10 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 118 + 2809422/1640-1760 ACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGU-CCAAAGGACAAUCCACAGCUUCGGUAAUAUGUUU-AG ..(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((-((...)))))))....))).))))).........-.. ( -22.70) >sy_ha.0 1800356 118 - 2685015/1640-1760 ACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGU-CCUACGGACAAUCCACAGCUUCGGUAAUAUGUUU-AG ..(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((-((...)))))))....))).))))).........-.. ( -22.70) >sp_ag.0 1990286 120 - 2160267/1640-1760 AUACCGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUGACACUUUUGUGUCAUCCACAGCUUCGGUAAUAUGUUUUAG .((((((.........((((((((((....)))).......((....))..........)))))).(((........(((((((....)))))))....))).))))))........... ( -25.70) >consensus ACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGU_CCUAAGGACAAUCCACAGCUUCGGUAAUAUGUUU_AG ..(((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........(((((........)))))....))).)))))............ (-17.76 = -15.10 + -2.66)

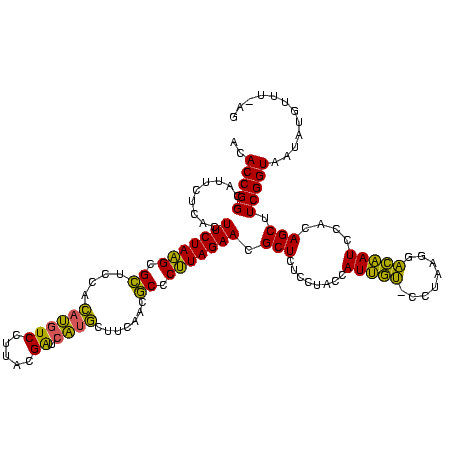
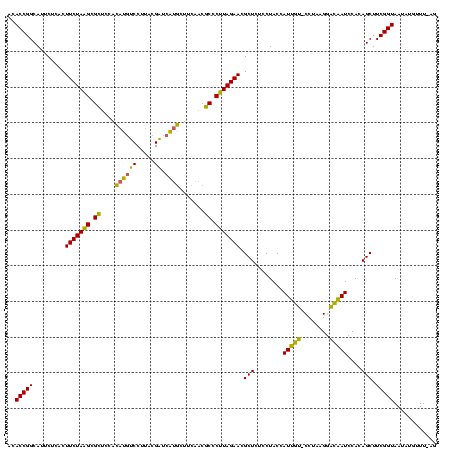
| Location | 2,228,674 – 2,228,792 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.84 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -38.33 |
| Energy contribution | -33.90 |
| Covariance contribution | -4.43 |
| Combinations/Pair | 1.34 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998398 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 118 + 2809422/1640-1760 CU-AAACAUAUUACCGAAGCUGUGGAUUGUCCUUUGG-ACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGU ..-........((((..........(((((((...))-)))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))))). ( -37.90) >sy_ha.0 1800356 118 - 2685015/1640-1760 CU-AAACAUAUUACCGAAGCUGUGGAUUGUCCGUAGG-ACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGU ..-........((((..........(((((((...))-)))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))))). ( -38.90) >sp_ag.0 1990286 120 - 2160267/1640-1760 CUAAAACAUAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAU ...........((((..........(((((((....)))))))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....)))))))). ( -43.90) >consensus CU_AAACAUAUUACCGAAGCUGUGGAUUGUCCAUAGG_ACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGU ...........((((..........(((((.(....).)))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))))). (-38.33 = -33.90 + -4.43) # Strand winner: reverse (1.00)

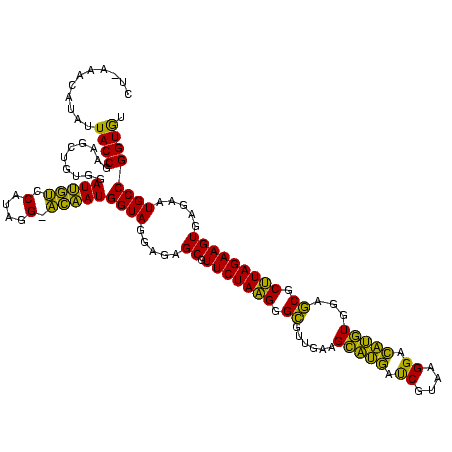
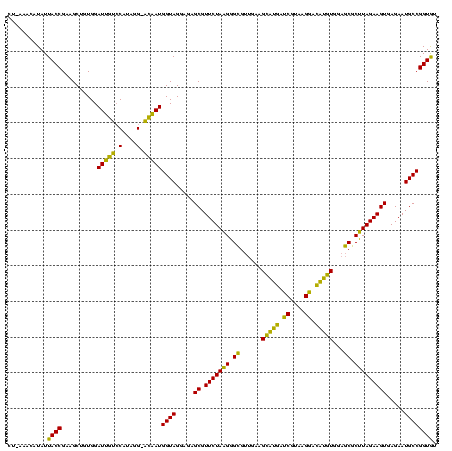
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -36.51 |
| Consensus MFE | -33.15 |
| Energy contribution | -31.17 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/1920-2040 UAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAA ........((((((((.((((....))))((((....)))).))))))))...(((((.(.((((..........)))).).)).(((((((((........))))))))).....))). ( -38.00) >sy_ha.0 1800356 120 - 2685015/1920-2040 UAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAA ........((((((((.((((....))))((((....)))).))))))))...(((((.(.((((..........)))).).)).(((((((((........))))))))).....))). ( -38.00) >sp_ag.0 1990286 120 - 2160267/1920-2040 UAGAGCACUGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAGAUAUAAUCGGCAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCGAAAGGGAA .........(((((..(((((((.....)))((....))...))))..)))))(((((.((((..............)))).)).(((((((..(......)..))))))).....))). ( -33.54) >consensus UAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAA ........((((((((...(((((((.....))))).))...))))))))...(((((.(.((((..........)))).).)).((((((((((......)))))))))).....))). (-33.15 = -31.17 + -1.99) # Strand winner: reverse (1.00)

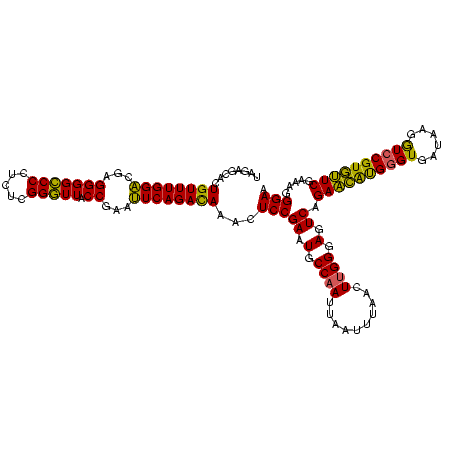
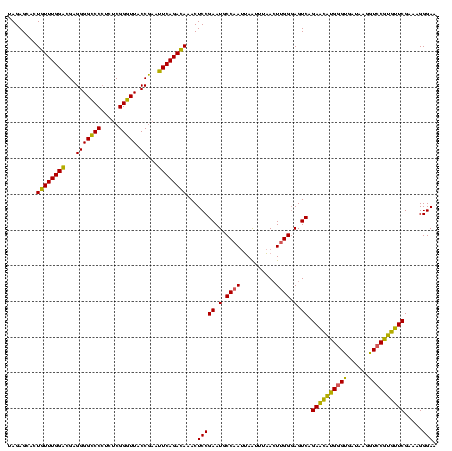
| Location | 2,228,674 – 2,228,793 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.96 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -35.86 |
| Energy contribution | -32.87 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937268 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 119 + 2809422/1960-2080 AUAGCUUUAGGGCUAGCCUCAA-GUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGG .(((((....)))))..(((((-((...(((((((((.....(((...((((((((.((((....))))((((....)))).))))))))..)))......)))).)))))..))))))) ( -40.20) >sy_ha.0 1800356 119 - 2685015/1960-2080 AUAGCUUUAGGGCUAGCCUCAA-GUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGG .(((((....)))))..(((((-((...(((((((((.....(((...((((((((.((((....))))((((....)))).))))))))..)))......)))).)))))..))))))) ( -40.20) >sp_ag.0 1990286 120 - 2160267/1960-2080 AUAGCUUUAGGGCUAGCGUCGAUGUUAAGUCUCUUGGAGGUAGAGCACUGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAGAUAUAAUCGGC .(((((....)))))..((((((.....(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))...)))))) ( -36.70) >consensus AUAGCUUUAGGGCUAGCCUCAA_GUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGG .(((((....)))))..(((((.((...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))) (-35.86 = -32.87 + -2.99) # Strand winner: reverse (1.00)

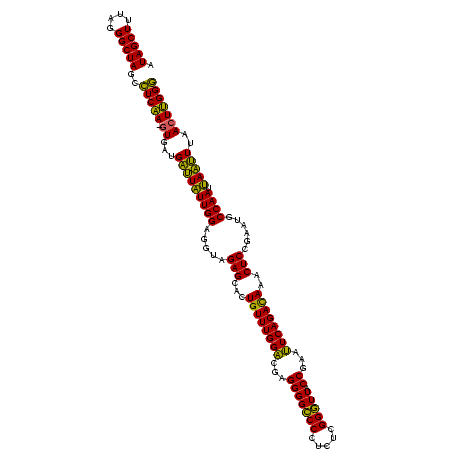
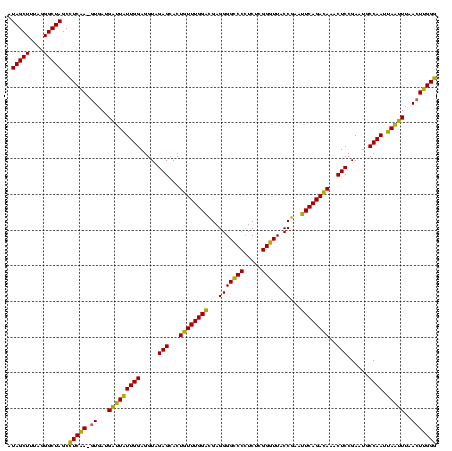
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -36.77 |
| Energy contribution | -34.67 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2080-2200 GUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAA ......((((((......))))))(((((((((..(((.(((((........)))))))).....((..((((.(.((((...))))...)..))))..)).....)))))))))..... ( -34.20) >sy_ha.0 1800356 120 - 2685015/2080-2200 GUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCUCUCCGAA ......((((((......))))))(((((((((..(((.(((((........)))))))).....((((.....)))).....((((((......)))))).....)))))))))..... ( -35.60) >sp_ag.0 1990286 120 - 2160267/2080-2200 GUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCCGAA ......((((((......))))))(((((...((((((((((((........)))))..........................((((((......))))))....))))))))))))... ( -37.00) >consensus GUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCUCUCCGAA ......((((((......))))))((((((((...(((.(((((........)))))))).............(((.......((((((......))))))...)))))))))))..... (-36.77 = -34.67 + -2.10) # Strand winner: reverse (1.00)

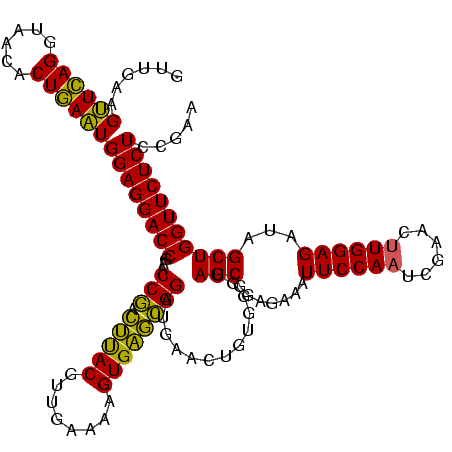

| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -27.04 |
| Energy contribution | -24.60 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2120-2240 CUCCGCUACCCUCAGUUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGACCAAGGGUAGAUCACCUGGUUUCGGGUCUACGACCA ...(((((((((..........(((.((........)).))).((((.(((......(((((......))))).....))).)))).)))))))...(((((....)))))...)).... ( -28.10) >sy_ha.0 1800356 120 - 2685015/2120-2240 CUCCGCUACCCACAGUUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGACCAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCA ...(((((((((..........(((.((........)).))).((((.(((......(((((......))))).....))).)))).)))))))...(((((....)))))...)).... ( -28.50) >sp_ag.0 1990286 120 - 2160267/2120-2240 CUCCGCUACCCACAAGUCAUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGAGUUUUACCUCACCUUCAACCUGCUCAUGGGUAGGUCACAUGGUUUCGGGUCUACAACAU .......((((.........(((.((((........))))..)))...((....((((((((......))))).....(((((((....)))))))....)))..))))))......... ( -30.30) >consensus CUCCGCUACCCACAGUUCAUCCGCUCACUUUUCAACGUAAGUCGGUUCGGUCCUCCAUUCAGUGUUACCUGAACUUCAACCUGACCAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCA .....(((((((..(((...(((((.((........)).)).)))...(((......(((((......))))).....))).)))..)))))))...(((((....)))))......... (-27.04 = -24.60 + -2.44)

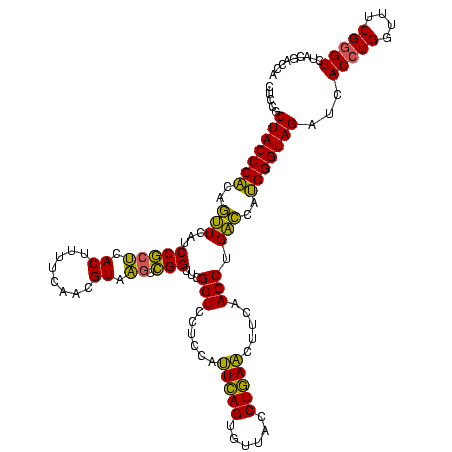
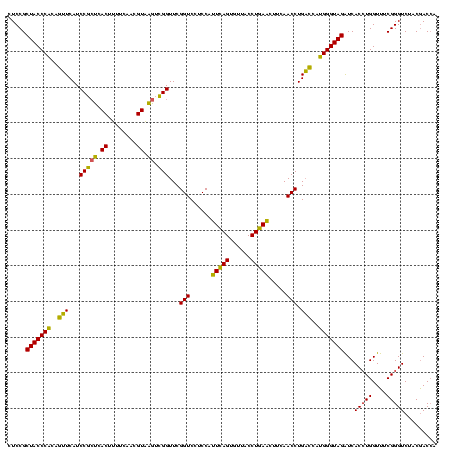
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.67 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -44.70 |
| Energy contribution | -40.93 |
| Covariance contribution | -3.77 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.49 |
| Structure conservation index | 1.06 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2120-2240 UGGUCGUAGACCCGAAACCAGGUGAUCUACCCUUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAG .((((...))))..............(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))..... ( -41.90) >sy_ha.0 1800356 120 - 2685015/2120-2240 UGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAG .((((...))))..............(((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)))))))))))..... ( -41.60) >sp_ag.0 1990286 120 - 2160267/2120-2240 AUGUUGUAGACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAG ...........(((....(....)..(((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..)))))))))).. ( -42.50) >consensus UGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAG ...........(((............(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..)))))))))).. (-44.70 = -40.93 + -3.77) # Strand winner: reverse (1.00)

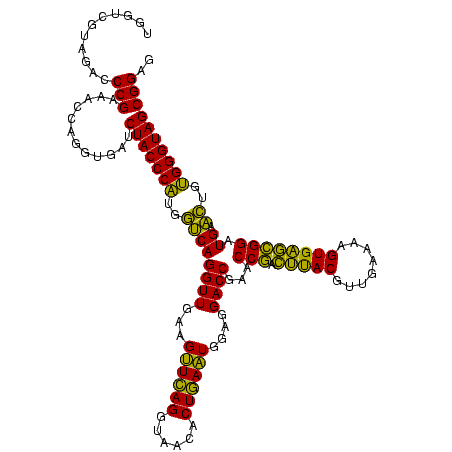
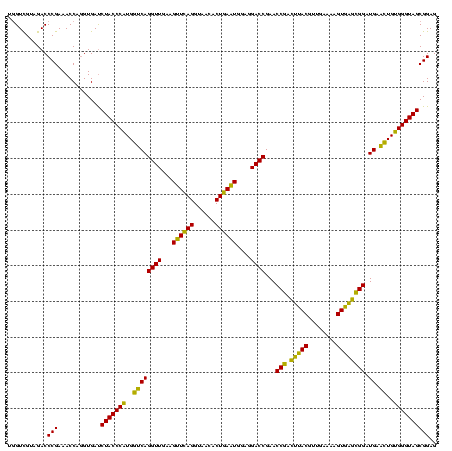
| Location | 2,228,674 – 2,228,793 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.07 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -34.86 |
| Energy contribution | -33.10 |
| Covariance contribution | -1.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.836385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 119 + 2809422/2200-2320 CGAGUUACGAUUUGAUGCAAGGUUAAGCAGUAAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU-UAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCUUGGUCAG ((.((((((((..(((((..((((..(((.....)))..))))...((....))..((((.....))))...-..)))))..))))).))).))..((((((.(......)))))))... ( -35.80) >sy_ha.0 1800356 119 - 2685015/2200-2320 CGAGUUACGAUCUGAUGCAAGGUUAAGCAGGAGAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU-GAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGUCAG ((.(((((((((.(((((..((((..(((.....)))..))))...((....))..((((.....))))...-..))))).)))))).))).))..((((((........)).))))... ( -35.80) >sp_ag.0 1990286 120 - 2160267/2200-2320 CGAGUUACGAUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUUAAUAGGGCGUCAUAGUAUCAUGUUGUAGACCCGAAACCAUGUGACCUACCCAUGAGCAG ((.(((((((((((((((..((((..(((.....)))..))))...((....))((((((.....)))).))...)))))))))))).))).))....((((.(......)))))..... ( -36.40) >consensus CGAGUUACGAUAUGAUGCAAGGUUAAGCAGAAGAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCGUU_UAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGUCAG ((.(((((((((.(((((..((((..(((.....)))..))))...((....))..((((.....))))......))))).)))))).))).))..((((((.(......)))))))... (-34.86 = -33.10 + -1.76) # Strand winner: reverse (1.00)

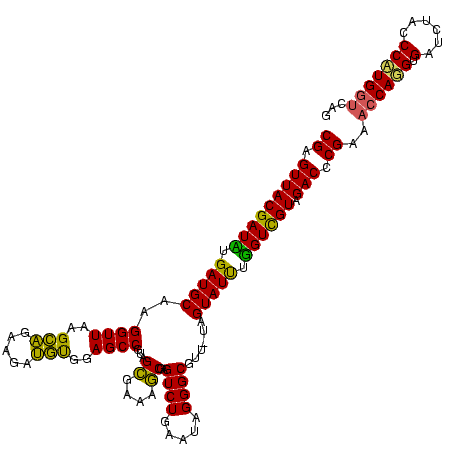
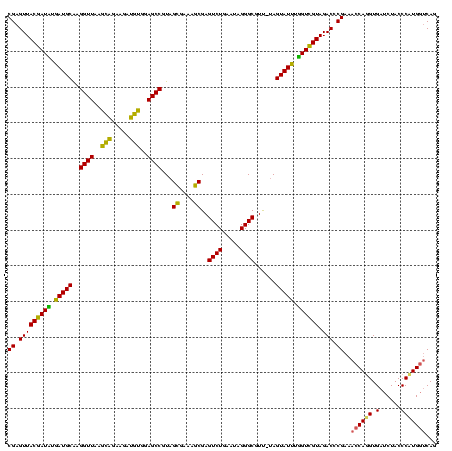
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -38.50 |
| Energy contribution | -37.29 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2320-2440 CCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGG ((....))...((((.....((.((((....)))).))........))))....(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....))))) ( -40.32) >sy_ha.0 1800356 120 - 2685015/2320-2440 CCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAGAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGG ((....))(((((((...(((((((((....)))((((........))))........)))))).......((((..(((((....)))))..))))..))))))).............. ( -39.50) >sp_ag.0 1990286 120 - 2160267/2320-2440 CCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAACCGG ((....))...((((.....((.((((....)))).))........))))....(((.((..((((((((.((((..(((((....)))))..)))).))....)))))).....))))) ( -39.72) >consensus CCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGG ((....))...((((.....((.((((....)))).))........))))....(((.((..(((((((..((((..(((((....)))))..))))......))))))).....))))) (-38.50 = -37.29 + -1.21) # Strand winner: reverse (1.00)

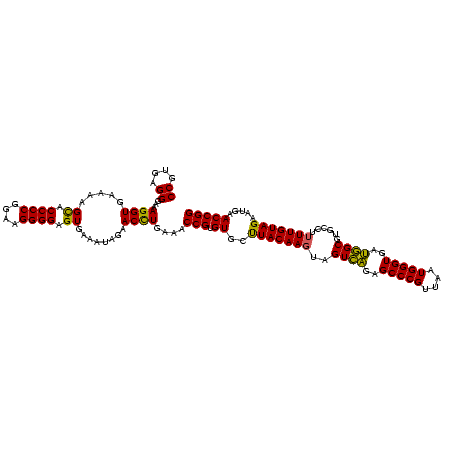
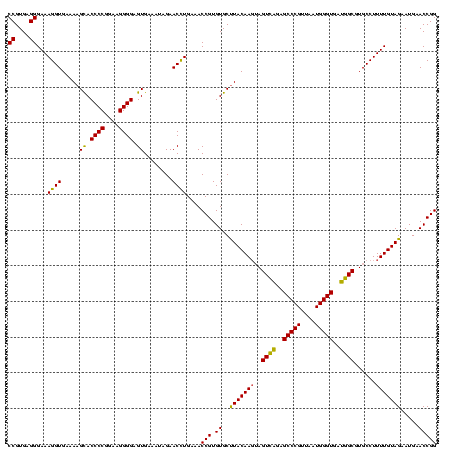
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -36.53 |
| Energy contribution | -35.87 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2400-2520 AGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGA ..(((((....(((.(((((....((((((......))))))..((((.........))))..))))).....(((....((....))....))))))..)).((((....)))).))). ( -37.10) >sy_ha.0 1800356 120 - 2685015/2400-2520 AGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGA ..(((......(((.(((((....((((((......))))))..((((.........))))..))))).....(((....((....))....)))))).....((((....)))).))). ( -36.50) >sp_ag.0 1990286 120 - 2160267/2400-2520 GACACGCGAAAUCUCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGA ..(((((....((.((((((....((((((......))))))........((((....)))).))))).).))(((....((....))....))).....)).((((....)))).))). ( -40.30) >consensus AGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGA ..(((((........(((((....((((((......))))))........((((....)))).))))).....(((....((....))....))).....)).((((....)))).))). (-36.53 = -35.87 + -0.66) # Strand winner: reverse (0.99)

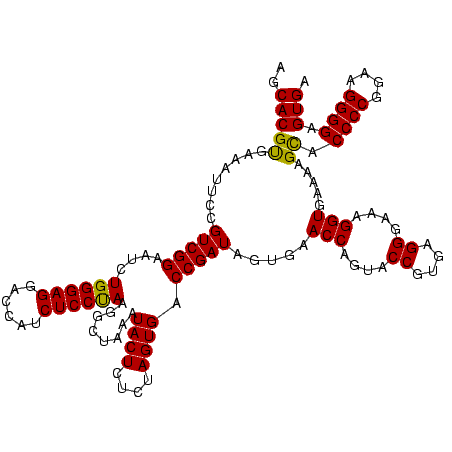
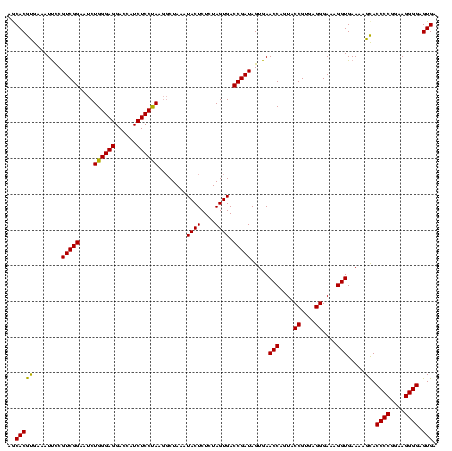
| Location | 2,228,674 – 2,228,793 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.19 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -19.53 |
| Energy contribution | -16.00 |
| Covariance contribution | -3.53 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 119 + 2809422/2520-2640 CUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUCUCUCUUGAGUGG-AUCCUGAGUACGACGG .......((..((.((..(((((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))).(((....)))...-.))))..))))..)). ( -33.20) >sy_ha.0 1800356 119 - 2685015/2520-2640 CUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGG-AUCCUGAGUACGACGG .......((..((.((..(((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...))))))).(((....)))...-.))))..))))..)). ( -23.80) >sp_ag.0 1990286 114 - 2160267/2520-2640 UGCGAUAUAGGAUUA------AUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGUACGGCGA (((.....(((.(((------((((((((....(..((((((....))))))..)...(((.((....))))).........))))).)))))))))..)))((((.....))))..... ( -23.90) >consensus CUCUAUACGGAGUUACAAAGGAACAUAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAGUCUCUCUUGAGUGG_AUCCUGAGUACGACGG .......(((..........(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))((....))........))).......... (-19.53 = -16.00 + -3.53) # Strand winner: reverse (1.00)

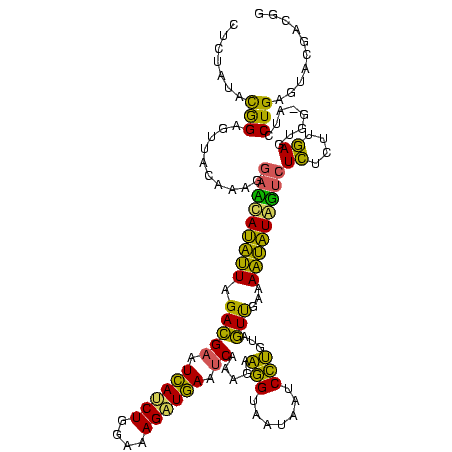
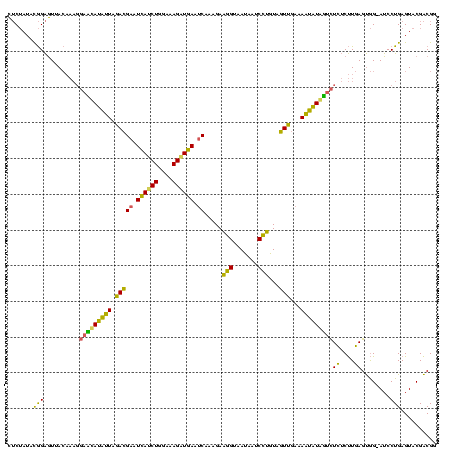
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -27.09 |
| Consensus MFE | -23.95 |
| Energy contribution | -21.30 |
| Covariance contribution | -2.65 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.92 |
| SVM RNA-class probability | 0.999961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2560-2680 CGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCU .(((...(((.......))).....((.((((((....)))))).)).......)))((((.(((((((((......))))....)))))..((((((((....))))))))...)))). ( -32.30) >sy_ha.0 1800356 120 - 2685015/2560-2680 CAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCU ....(((((((.(((.........(((.(((.((....)).))).)))......))).)))..))))((((......))))((((.......((((((((....))))))))...)))). ( -23.76) >sp_ag.0 1990286 113 - 2160267/2560-2680 UAAAUACGAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCU ........(((((((..........(..((((((....))))))..).........(((------.........))))))-).)))......((((((((....))))))))........ ( -25.20) >consensus CAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAAUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUUAUUGGUUUGGGCU .......(((((.............((.((((((....)))))).))....................((((......))))))))).(((..((((((((....)))))))).))).... (-23.95 = -21.30 + -2.65)

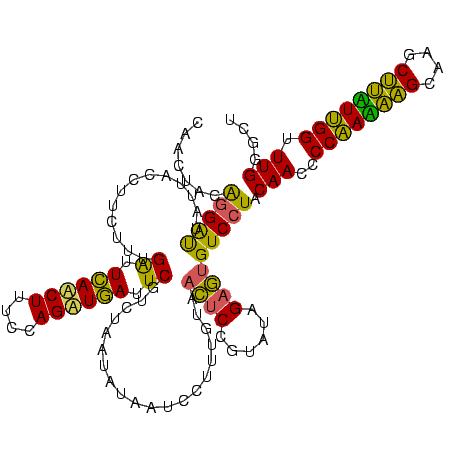
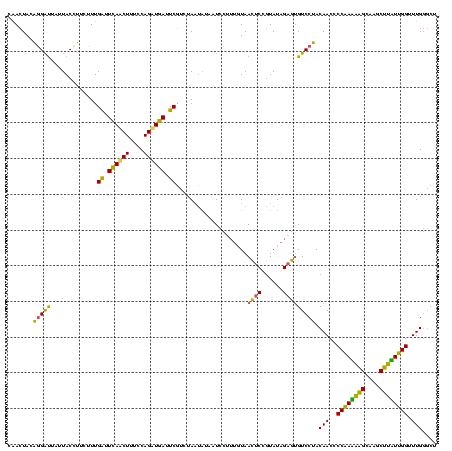
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -27.23 |
| Energy contribution | -24.80 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.90 |
| SVM RNA-class probability | 0.999960 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
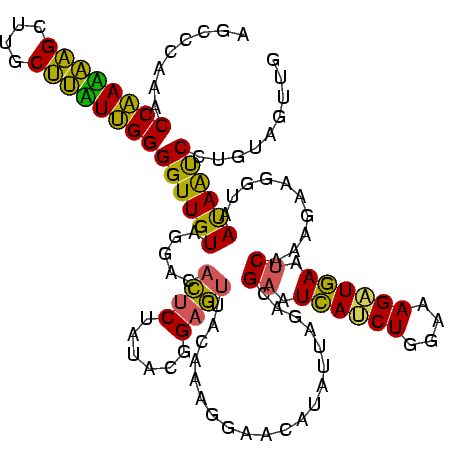
>sy_au.0 2228674 120 + 2809422/2560-2680 AGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCG ...((...((((((((....))))))))..(((((....(((((....)))))))))).)).((((.......((.((((((....)))))).)).....(((.......)))...)))) ( -36.50) >sy_ha.0 1800356 120 - 2685015/2560-2680 AGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUG ...((...((((((((....))))))))..(((((....(((((....)))))))))).)).........(((((.(((.((....)).))).)).....(((.......)))...))). ( -26.60) >sp_ag.0 1990286 113 - 2160267/2560-2680 AGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUA------AUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUA ..(((...((((((((....)))))))).((((((....-)))))).........------...............((((((....)))))))))...(((.((....)))))....... ( -32.10) >consensus AGCCCAAACCAAAAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACAUAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUG ........((((((((....))))))))((((((.....((((......))))....................((.((((((....)))))).))..........))))))......... (-27.23 = -24.80 + -2.43) # Strand winner: reverse (0.96)


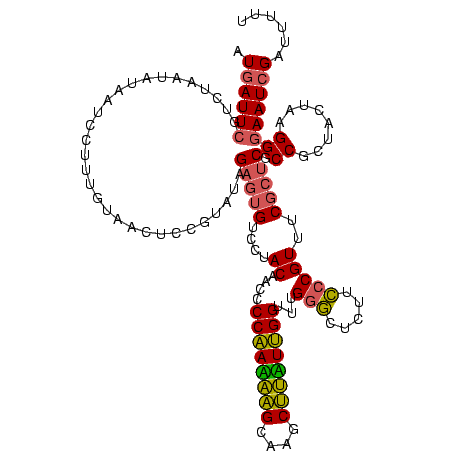
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -21.54 |
| Energy contribution | -21.10 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2600-2720 AUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUU ..((((((.........((((.(((((((((......))))....)))))..((((((((....))))))))...))))..(((((.....((.....))......))))).)))))).. ( -32.40) >sy_ha.0 1800356 120 - 2685015/2600-2720 AUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUU .((((((..............................(((((....((....((((((((....))))))))...(((.....)))))..))))).((........))))))))...... ( -27.50) >sp_ag.0 1990286 113 - 2160267/2600-2720 GUAAUUCUUCUAUAAUGAU------UAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUG ................(((------(...........(((-(.....(((..((((((((....)))))))).))).....))))..........(((........)))))))....... ( -25.20) >consensus AUGAUUCGUCUAAUAUAAUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAAAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAUUUUU .((((((..............................(((((....((....((((((((....))))))))...(((.....)))))..))))).((........))))))))...... (-21.54 = -21.10 + -0.44)

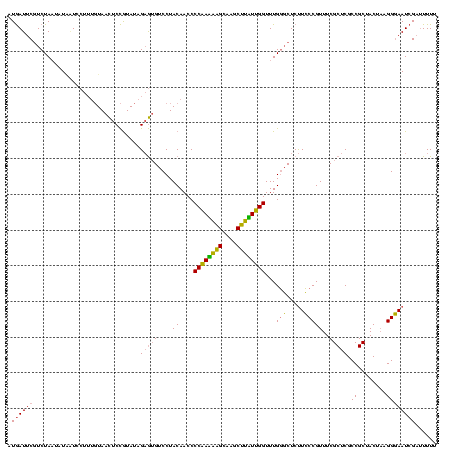
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -25.54 |
| Energy contribution | -23.77 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2600-2720 AAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAU .......(((((((((.(((((..(((((.....(((.....)))...((((((((....)))))))).)))(((((....))))).))..))))).)))).(.......)..))))).. ( -34.70) >sy_ha.0 1800356 120 - 2685015/2600-2720 AAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAU .......((((((((..(((((..(((((.....(((.....)))...((((((((....)))))))).)))(((((....))))).))..)))))..)))............))))).. ( -31.10) >sp_ag.0 1990286 113 - 2160267/2600-2720 CAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUA------AUCAUUAUAGAAGAAUUAC ......((.(((((........))))).)).....((((....((((.((((((((....))))))))..))))....)-))).......((((.------.((......))..)))).. ( -33.00) >consensus AAAAAUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAAAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACAUAUUAGACGAAUCAU .......((((((((.....(((((.........(((.....)))...((((((((....)))))))).))))))))..((((......))))....................))))).. (-25.54 = -23.77 + -1.77) # Strand winner: reverse (0.99)

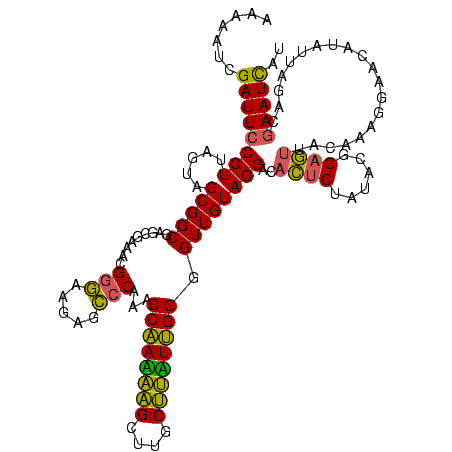
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.00 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -30.29 |
| Energy contribution | -26.97 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2640-2760 CGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACA (((.(.(((((......))))).))))..((((..(((......)))..))))....((....((((.(.....(((.....)))...((((((((....))))))))).))))...)). ( -35.10) >sy_ha.0 1800356 120 - 2685015/2640-2760 CGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACA (((.(.(((((......))))).))))..((((..((........))..))))....((....((((.(.....(((.....)))...((((((((....))))))))).))))...)). ( -32.10) >sp_ag.0 1990286 119 - 2160267/2640-2760 CAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC- (((....)))...(.(((..((.((((((((....((..((....))..)).)))..))))).))..((......)).))).)((((.((((((((....))))))))..)))).....- ( -34.50) >consensus CGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAAAAGCUUGCUUAUUGGGGUUGUAGGACA (((....))).....(((((..((((((...........((....))..(((((......((.....)).....)))))....))....(((((((....)))))))))))..))))).. (-30.29 = -26.97 + -3.32) # Strand winner: reverse (1.00)

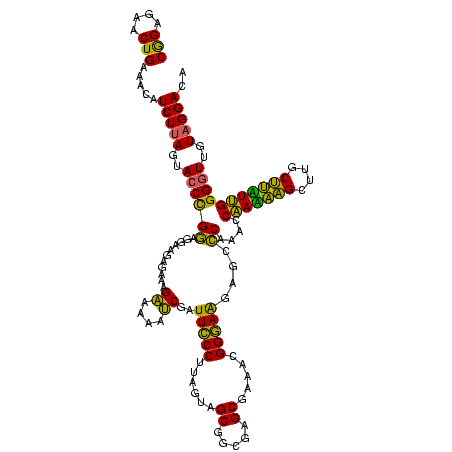
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -22.44 |
| Energy contribution | -19.33 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.28 |
| Mean z-score | -0.19 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2680-2800 CUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUGUGCCUUCUGAUAUGCUAUGUAUUCACAUAUCGAUAA ...........((.((((........(((((..(((......)))..))))).((((.(((..((....))))).).))))))).))....(((((((...........))))))).... ( -23.80) >sy_ha.0 1800356 120 - 2685015/2680-2800 CUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUCUGCCUUCUGACAAGCUAUGAAUUCACUUGUCGAUAA ..((((.....((.....))......))))((((((......(((.......(((((.(((..((....))))).).))))(((........))).......))).......)))))).. ( -23.92) >sp_ag.0 1990286 116 - 2160267/2680-2800 CCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAA----CAGAUAUGGAUAC .((((..........(((........)))....((....))...........))))......(((((...(((....))))))))....((.((((((.....----..))))))))... ( -20.80) >consensus CUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAUUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUCUGCCUUCUGAUAUGCUAUG_AUUCACAUAUCGAUAA ..((((.....((.....))......))))(((..................(((((..(((..((....)))))...)))))..........((((((...........))))))))).. (-22.44 = -19.33 + -3.10)

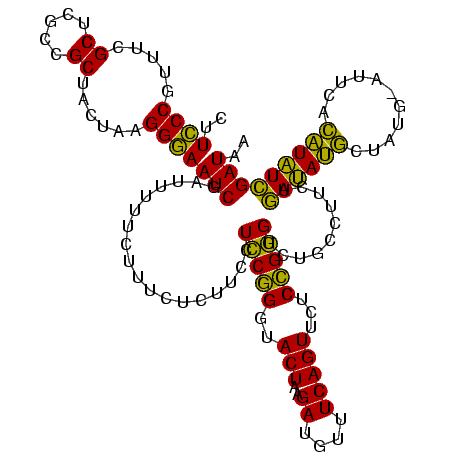

| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -26.80 |
| Energy contribution | -23.37 |
| Covariance contribution | -3.43 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2680-2800 UUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAG ...(((((((((.........)))))))........(..((((.(.(((((......))))).)))))..)..))(((......)))..(((((......((.....)).....))))). ( -27.50) >sy_ha.0 1800356 120 - 2685015/2680-2800 UUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAG ...(((....(((....))).(((((((......(((..((((.(.(((((......))))).))))).((((..((........))..)))))))......)))))))....))).... ( -30.90) >sp_ag.0 1990286 116 - 2160267/2680-2800 GUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGG ...(((((((((.----....))))))).))........(((((..(((..........))).)))))...........((.....((.(((((........))))).)).....))... ( -28.10) >consensus UUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAG ...(((((((((.........))))))).))........((((...(((((......)))))..))))...........((....))..(((((......((.....)).....))))). (-26.80 = -23.37 + -3.43) # Strand winner: reverse (0.98)

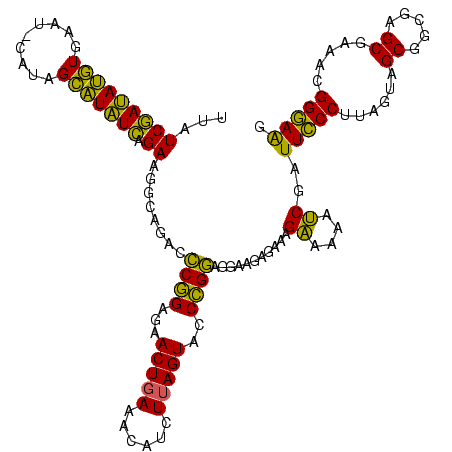
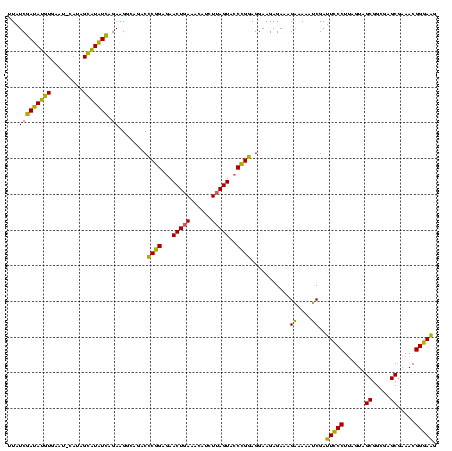
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -25.03 |
| Consensus MFE | -21.19 |
| Energy contribution | -18.53 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2720-2840 CUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUGUGCCUUCUGAUAUGCUAUGUAUUCACAUAUCGAUAACAUGACAUAACUCAUGCUGGGUUUCCCCAUUCGGAAAUCU ...........(((((((.....((....))........(((...((((..(((((((...........)))))))....(((((......)))))..))))...))))))))))..... ( -25.00) >sy_ha.0 1800356 120 - 2685015/2720-2840 CUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUCUGCCUUCUGACAAGCUAUGAAUUCACUUGUCGAUAACACAACAUAACUCGUGCUGGGUUCCCCCAUUCGGAAAUCU ...........(((((((((....(((((...(((.((.(((....)))...((((((...((....)))))))))).)))..))))).....)))))(((....)))...))))..... ( -26.70) >sp_ag.0 1990286 113 - 2160267/2720-2840 CUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAA----CAGAUAUGGAUACUAGUCAUUAACUAG---UGGGUUCCCCCAUUCGGACAUCU ............((((..(((..((....)))))...))))((((....((.((((((.....----..)))))))).((((((.....)))))---)(((....)))....)))).... ( -23.40) >consensus CUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUCUGCCUUCUGAUAUGCUAUG_AUUCACAUAUCGAUAACACAACAUAACUCGUGCUGGGUUCCCCCAUUCGGAAAUCU ........((((((((..(((..((....)))))...))))).........(((((((...........))))))).....................((((....))))...)))..... (-21.19 = -18.53 + -2.66)

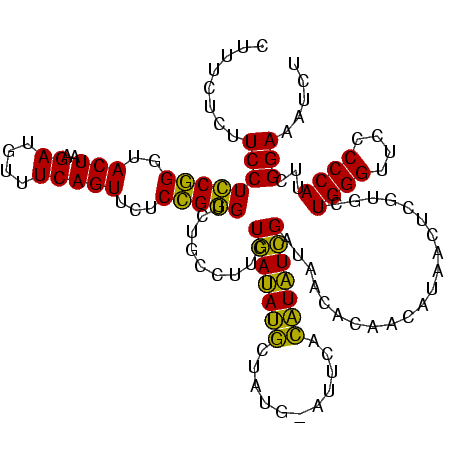
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.06 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -30.16 |
| Energy contribution | -28.83 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.57 |
| SVM RNA-class probability | 0.999400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2720-2840 AGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAG ...(((((((..(((....)))(((((((......))))))).))(((((((.........)))))))...........((((.(.(((((......))))).))))).)))))...... ( -36.70) >sy_ha.0 1800356 120 - 2685015/2720-2840 AGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUUGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAG ...(((((((..(((....)))(((((((......))))))).))(((((((.........)))))))...........((((.(.(((((......))))).))))).)))))...... ( -36.20) >sp_ag.0 1990286 113 - 2160267/2720-2840 AGAUGUCCGAAUGGGGGAACCCA---CUAGUUAAUGACUAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAG .....(((....(((....)))(---((((((...))))))).(((((((((.----....))))))).)).)))....(((((..(((..........))).)))))............ ( -31.80) >consensus AGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUAAUGUUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAG ...(((((...((((....))))...............((((.(((((((((.........))))))).)).))))...((((...(((((......)))))..)))).)))))...... (-30.16 = -28.83 + -1.33) # Strand winner: reverse (1.00)

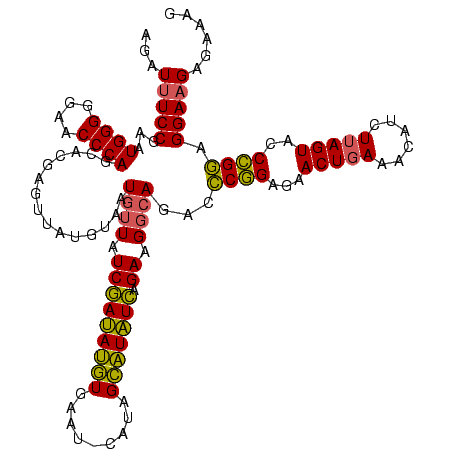
| Location | 2,228,674 – 2,228,794 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -29.24 |
| Energy contribution | -28.13 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 2228674 120 + 2809422/2760-2880 ACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACC ..((((((((((((((((((.......)))))...))))))......(((..(((....)))(((((((......))))))).))).(((((....))))))))))))............ ( -37.50) >sy_ha.0 1800356 120 - 2685015/2760-2880 ACGAUAUGCUUUGGGUAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUUGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACC ((((((((((((((((.(((.......)))....)))))))).....((..((((....))))...))...))))))))((..(((((((((.........))))))).))..))..... ( -37.70) >sp_ag.0 1990286 113 - 2160267/2760-2880 ACGAAAUGCUUUGGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCA---CUAGUUAAUGACUAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACG ........(((((((.(((.((......)))))..)))))))...(((....(((....)))(---((((((...))))))).(((((((((.----....))))))).)).)))..... ( -34.60) >consensus ACGAUAUGCUUUGGGGAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUAAUGUUAUCGAUAUGUGAAU_CAUAGCAUAUCAGAAGGCAGACC .((.....(((((((..(((.......))).....))))))).....))..((((....))))...............((((.(((((((((.........))))))).)).)))).... (-29.24 = -28.13 + -1.11) # Strand winner: reverse (1.00)

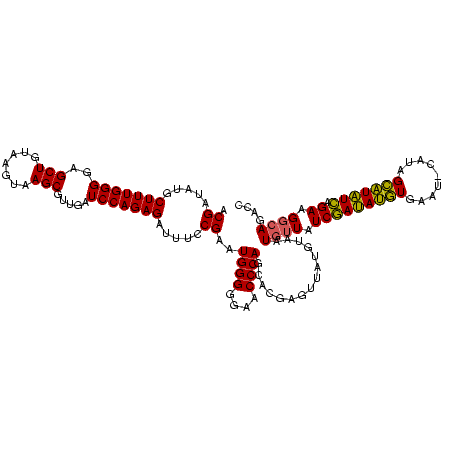
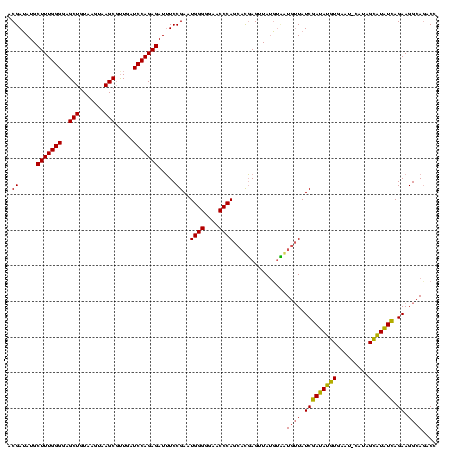
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:58:38 2006