
| Sequence ID | nosto.0 |
|---|---|
| Location | 1,819,469 – 1,819,685 |
| Length | 216 |
| Max. P | 0.998677 |

| Location | 1,819,469 – 1,819,588 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.31 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -32.31 |
| Energy contribution | -30.15 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
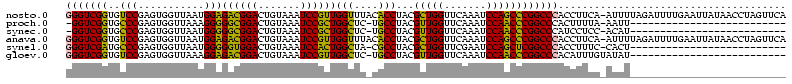
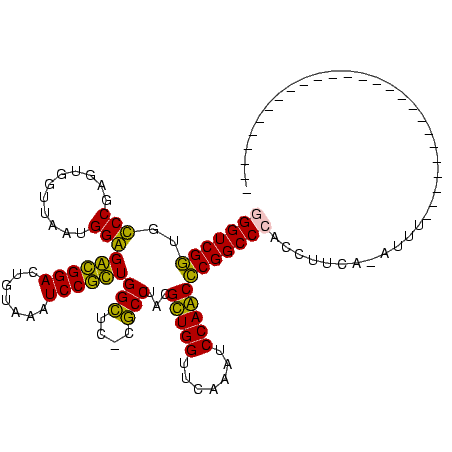
>nosto.0 1819469 119 + 6413771 GGGUCGGUGUCCGAGUGGUUAAUGGAGACGGACUGUAAAUCCGUUGGUUUACACCUACGCUGGUUCAAAUCCAGCCCGGCCCACCUUCA-AUUUUAGAUUUUGAAUUAUAACCUAGUUCA (((((((.((((..((.....))(....)))))((((((((....)))))))).....(((((.......)))))))))))).......-...........(((((((.....))))))) ( -35.90) >proch.0 1343538 91 - 1709204 -GGUCGGUGCCCGAGUGGUUAAAGGGGGCGGACUGUAAAUCCGCUGGCUC-UGCCUACGUUGGUUCAAAUCCAACCCGGCCCACUUUUA-AAUU-------------------------- -((((((.......((((....((((((((((.......))))))..)))-)..))))(((((.......)))))))))))........-....-------------------------- ( -30.10) >synec.0 1843149 91 + 2434428 -GGUCGGUGCCCGAGUGGUUAAUGGGGGCGGACUGUAAAUCCGCUGGCUC-UGCCUACGUUGGUUCAAAUCCAACCCGGCCCAUCCUCC-ACAU-------------------------- -..((((...))))((((...(((((((((((.......))))))(((..-.)))...(((((.......)))))....)))))...))-))..-------------------------- ( -32.50) >anava.0 6278818 119 + 6365727 GGGUCGGUGUCCGAGUGGUUAAUGGAGACGGACUGUAAAUCCGUUGGUUUACACCUACGCUGGUUCAAAUCCAGCCCGGCCCACCUUCA-AUUUUAGAUUUUGAAUUAUAACCUAGUUCA (((((((.((((..((.....))(....)))))((((((((....)))))))).....(((((.......)))))))))))).......-...........(((((((.....))))))) ( -35.90) >synel.0 1998599 92 - 2696255 GGGUCGAUGCCCGAGUGGUUAAUGGGGGUGGACUGUAAAUCCACUGGCUA-CGCCUACGCUGGUUCGAAUCCAGCUCGGCCCACCUUUC-CACU-------------------------- ((((....)))).(((((....((((((((((.......))))))(((..-.)))...(((((.......)))))....)))).....)-))))-------------------------- ( -34.20) >gloev.0 3245929 93 - 4659019 GGGUCGGUGUCCGAGUGGUUAAAGGAGACGGACUGUAAAUCCGUUGGCUC-UGCCUACGUUGGUUCAAAUCCAACCCGGCCCACAUUUGUAUAU-------------------------- (((((((.......((((.((.((..((((((.......))))))..)).-)).))))(((((.......))))))))))))............-------------------------- ( -32.10) >consensus GGGUCGGUGCCCGAGUGGUUAAUGGAGACGGACUGUAAAUCCGCUGGCUC_CGCCUACGCUGGUUCAAAUCCAACCCGGCCCACCUUCA_AUUU__________________________ (((((((..(((...........)))((((((.......))))))(((....)))...(((((.......))))))))))))...................................... (-32.31 = -30.15 + -2.16)
| Location | 1,819,469 – 1,819,588 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.31 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -28.58 |
| Energy contribution | -26.42 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
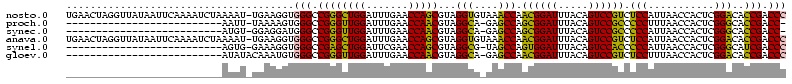
>nosto.0 1819469 119 - 6413771 UGAACUAGGUUAUAAUUCAAAAUCUAAAAU-UGAAGGUGGGCCGGGCUGGAUUUGAACCAGCGUAGGUGUAAACCAACGGAUUUACAGUCCGUCUCCAUUAACCACUCGGACACCGACCC .......((((....(((((.........)-))))((((..(((((((((.......)))))((.(((........((((((.....))))))........))))))))).)))))))). ( -35.29) >proch.0 1343538 91 + 1709204 --------------------------AAUU-UAAAAGUGGGCCGGGUUGGAUUUGAACCAACGUAGGCA-GAGCCAGCGGAUUUACAGUCCGCCCCCUUUAACCACUCGGGCACCGACC- --------------------------....-.....(((((((..(((((.......)))))...)))(-(((...((((((.....))))))...))))..))))((((...))))..- ( -30.00) >synec.0 1843149 91 - 2434428 --------------------------AUGU-GGAGGAUGGGCCGGGUUGGAUUUGAACCAACGUAGGCA-GAGCCAGCGGAUUUACAGUCCGCCCCCAUUAACCACUCGGGCACCGACC- --------------------------..((-((..((((((((..(((((.......)))))...))).-......((((((.....))))))..)))))..))))((((...))))..- ( -34.80) >anava.0 6278818 119 - 6365727 UGAACUAGGUUAUAAUUCAAAAUCUAAAAU-UGAAGGUGGGCCGGGCUGGAUUUGAACCAGCGUAGGUGUAAACCAACGGAUUUACAGUCCGUCUCCAUUAACCACUCGGACACCGACCC .......((((....(((((.........)-))))((((..(((((((((.......)))))((.(((........((((((.....))))))........))))))))).)))))))). ( -35.29) >synel.0 1998599 92 + 2696255 --------------------------AGUG-GAAAGGUGGGCCGAGCUGGAUUCGAACCAGCGUAGGCG-UAGCCAGUGGAUUUACAGUCCACCCCCAUUAACCACUCGGGCAUCGACCC --------------------------((((-(...((((((((..(((((.......)))))...))).-......((((((.....))))))..)))))..))))).(((......))) ( -35.00) >gloev.0 3245929 93 + 4659019 --------------------------AUAUACAAAUGUGGGCCGGGUUGGAUUUGAACCAACGUAGGCA-GAGCCAACGGAUUUACAGUCCGUCUCCUUUAACCACUCGGACACCGACCC --------------------------..........(((((((..(((((.......)))))...))).-(((...((((((.....)))))))))......))))((((...))))... ( -27.90) >consensus __________________________AAAU_UAAAGGUGGGCCGGGCUGGAUUUGAACCAACGUAGGCA_GAGCCAACGGAUUUACAGUCCGCCCCCAUUAACCACUCGGACACCGACCC ......................................(((.((((((((.......)))))...(((....))).((((((.....)))))).(((...........)))..))).))) (-28.58 = -26.42 + -2.16)
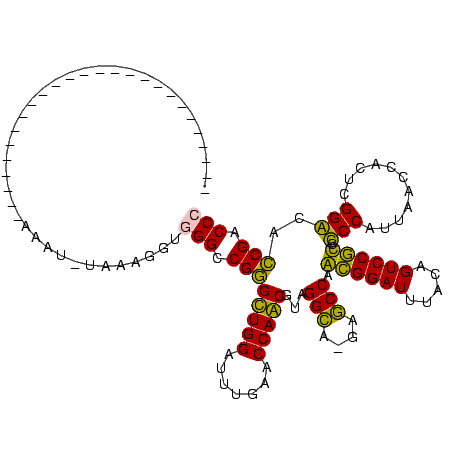
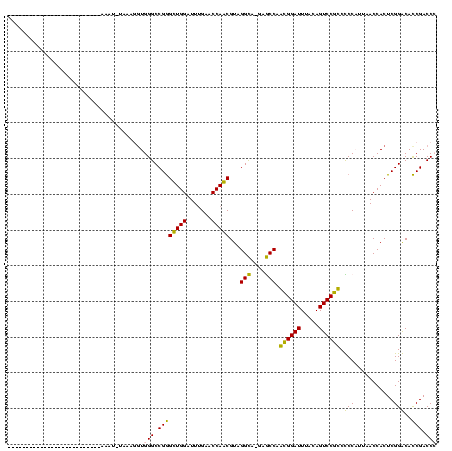
| Location | 1,819,509 – 1,819,628 |
|---|---|
| Length | 119 |
| Sequences | 2 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -34.65 |
| Energy contribution | -34.65 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 1819509 119 + 6413771 CCGUUGGUUUACACCUACGCUGGUUCAAAUCCAGCCCGGCCCACCUUCAAUUUUAGAUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAAAUUUCGCCCGUGUGGCUCAGU ...(((..((((((....(((((.......)))))..(((........(((((((((((((((((((.....)))))))))))))))))))............))).))))))..))). ( -34.65) >anava.0 6278858 119 + 6365727 CCGUUGGUUUACACCUACGCUGGUUCAAAUCCAGCCCGGCCCACCUUCAAUUUUAGAUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAACUUCCGCCCGUGUGGCUCAGU ...(((..((((((....(((((.......))))).(((.........(((((((((((((((((((.....)))))))))))))))))))..........)))...))))))..))). ( -34.81) >consensus CCGUUGGUUUACACCUACGCUGGUUCAAAUCCAGCCCGGCCCACCUUCAAUUUUAGAUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAAAUUCCGCCCGUGUGGCUCAGU ...(((..((((((....(((((.......)))))..(((........(((((((((((((((((((.....)))))))))))))))))))............))).))))))..))). (-34.65 = -34.65 + 0.00)

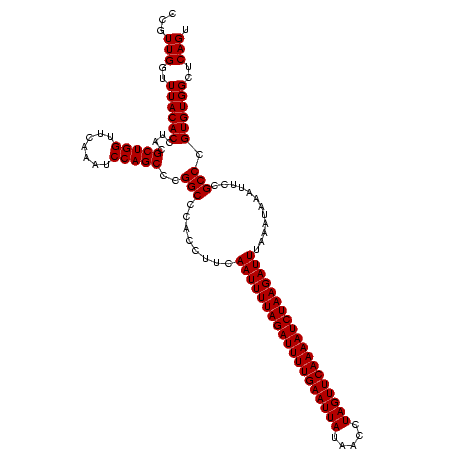
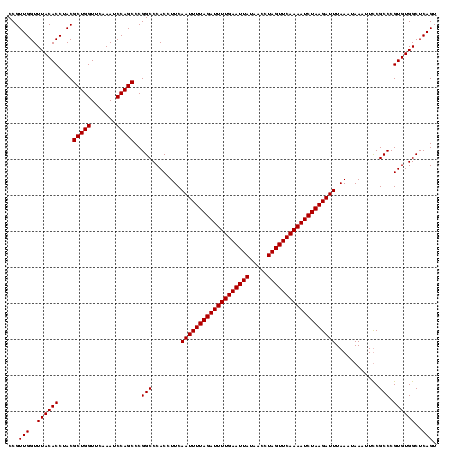
| Location | 1,819,509 – 1,819,628 |
|---|---|
| Length | 119 |
| Sequences | 2 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -35.45 |
| Energy contribution | -35.95 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994029 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 1819509 119 - 6413771 ACUGAGCCACACGGGCGAAAUUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAUCUAAAAUUGAAGGUGGGCCGGGCUGGAUUUGAACCAGCGUAGGUGUAAACCAACGG .(((.(((.(((...(((.((((....)))).((((((((((((.((.....)).))))))))))))..)))...)))))))))(((((.......)))))((.(((....))).)).. ( -34.40) >anava.0 6278858 119 - 6365727 ACUGAGCCACACGGGCGGAAGUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAUCUAAAAUUGAAGGUGGGCCGGGCUGGAUUUGAACCAGCGUAGGUGUAAACCAACGG .(((.(((.(((.(((....))).(((((...((((((((((((.((.....)).))))))))))))..))))).)))))))))(((((.......)))))((.(((....))).)).. ( -39.80) >consensus ACUGAGCCACACGGGCGAAAGUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAUCUAAAAUUGAAGGUGGGCCGGGCUGGAUUUGAACCAGCGUAGGUGUAAACCAACGG .(((.(((.(((.(((....))).(((((...((((((((((((.((.....)).))))))))))))..))))).)))))))))(((((.......)))))((.(((....))).)).. (-35.45 = -35.95 + 0.50)

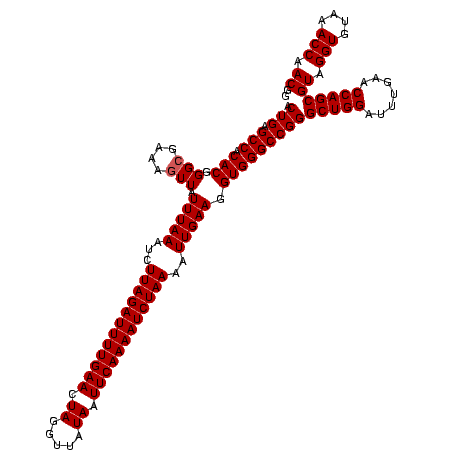
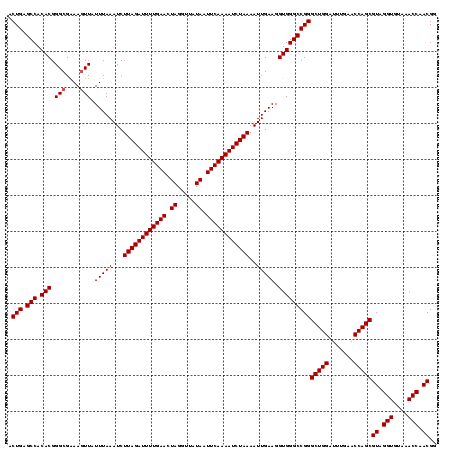
| Location | 1,819,549 – 1,819,668 |
|---|---|
| Length | 119 |
| Sequences | 2 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -38.40 |
| Energy contribution | -38.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.96 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 1819549 119 + 6413771 CCACCUUCAAUUUUAGAUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAAAUUUCGCCCGUGUGGCUCAGUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCACGAGUUCA .....((((((((((((((((((((((.....))))))))))))))))))).......((((((((((((((.(((.......))))))))(....)...)))))))))...))).... ( -38.50) >anava.0 6278898 119 + 6365727 CCACCUUCAAUUUUAGAUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAACUUCCGCCCGUGUGGCUCAGUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCACGAGUUCA ((((....(((((((((((((((((((.....)))))))))))))))))))............(((.....)))...))))..((((.((((((.....)))))).(....)..)))). ( -38.40) >consensus CCACCUUCAAUUUUAGAUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAAAUUCCGCCCGUGUGGCUCAGUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCACGAGUUCA ((((....(((((((((((((((((((.....)))))))))))))))))))............(((.....)))...))))..((((.((((((.....)))))).(....)..)))). (-38.40 = -38.40 + 0.00)

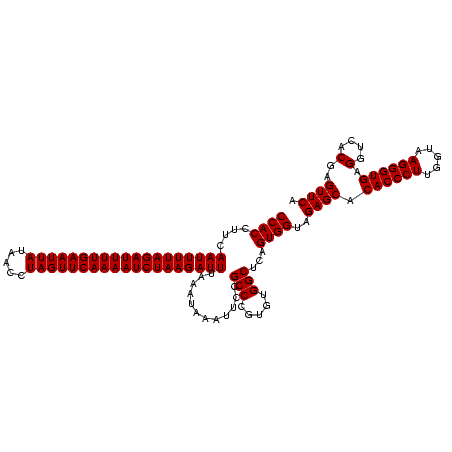
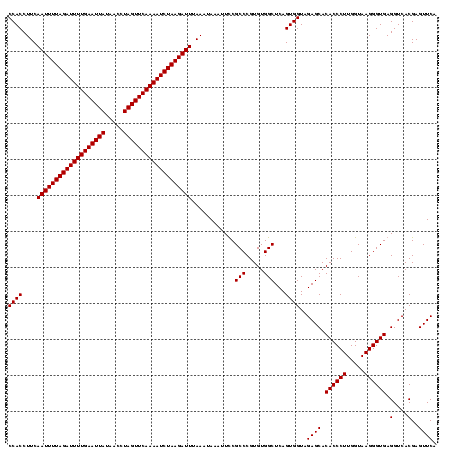
| Location | 1,819,549 – 1,819,668 |
|---|---|
| Length | 119 |
| Sequences | 2 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 98.32 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -34.20 |
| Energy contribution | -34.20 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 1819549 119 - 6413771 UGAACUCGUGACCUCACCCUUACCAAGGGUGUGCUCUACCACUGAGCCACACGGGCGAAAUUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAUCUAAAAUUGAAGGUGG ....((((((....((((((.....)))))).((((.......))))..)))))).......(((((.(((.((((((((((((.((.....)).)))))))))))).)))..))))). ( -34.20) >anava.0 6278898 119 - 6365727 UGAACUCGUGACCUCACCCUUACCAAGGGUGUGCUCUACCACUGAGCCACACGGGCGGAAGUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAUCUAAAAUUGAAGGUGG ....((((((....((((((.....)))))).((((.......))))..))))))(....).(((((.(((.((((((((((((.((.....)).)))))))))))).)))..))))). ( -34.60) >consensus UGAACUCGUGACCUCACCCUUACCAAGGGUGUGCUCUACCACUGAGCCACACGGGCGAAAGUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAUCUAAAAUUGAAGGUGG ....((((((....((((((.....)))))).((((.......))))..)))))).......(((((.(((.((((((((((((.((.....)).)))))))))))).)))..))))). (-34.20 = -34.20 + 0.00)

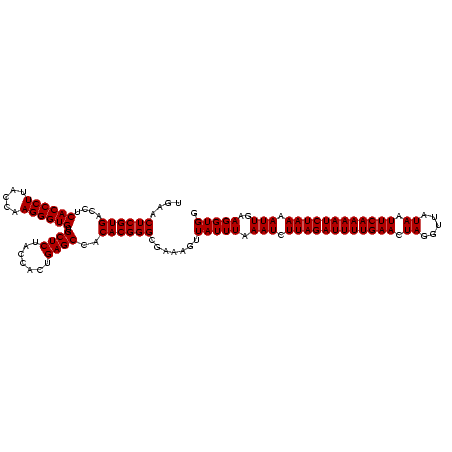
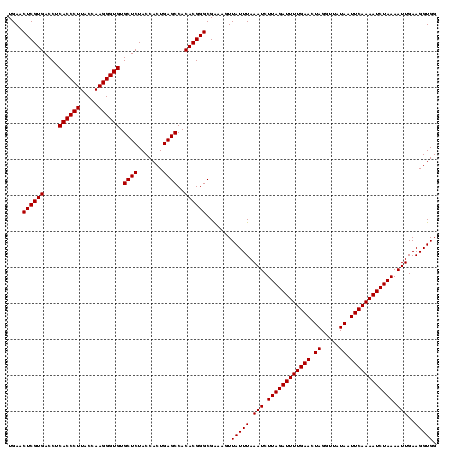
| Location | 1,819,565 – 1,819,685 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -39.30 |
| Energy contribution | -39.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 1819565 120 + 6413771 AUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAAAUUUCGCCCGUGUGGCUCAGUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCACGAGUUCAAUCCUCGUCACGGGCUU (((((((((((.....)))))))))))....................(((((((..((((.......)))).((((((.....)))))).....(((((.......)))))))))))).. ( -39.30) >anava.0 6278914 120 + 6365727 AUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAACUUCCGCCCGUGUGGCUCAGUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCACGAGUUCAAUCCUCGUCACGGGCUU (((((((((((.....)))))))))))....................(((((((..((((.......)))).((((((.....)))))).....(((((.......)))))))))))).. ( -39.30) >consensus AUUUUGAAUUAUAACCUAGUUCAAAAUCUAAGAUUUAAAUAAAUUCCGCCCGUGUGGCUCAGUGGUAGAGCACACCCUUGGUAAGGGUGAGGUCACGAGUUCAAUCCUCGUCACGGGCUU (((((((((((.....)))))))))))....................(((((((..((((.......)))).((((((.....)))))).....(((((.......)))))))))))).. (-39.30 = -39.30 + 0.00)

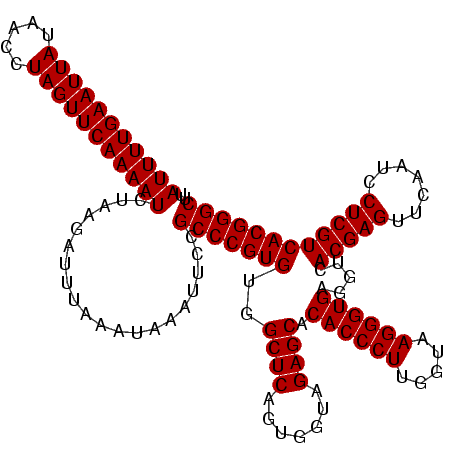
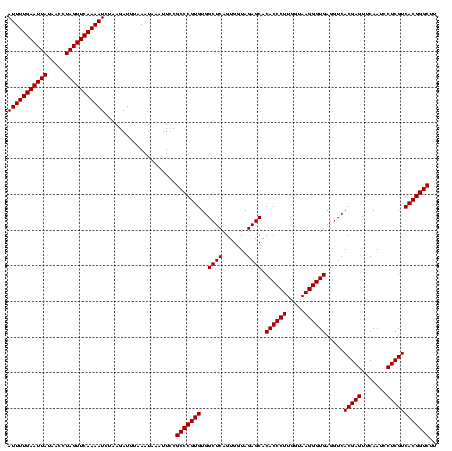
| Location | 1,819,565 – 1,819,685 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -38.80 |
| Energy contribution | -38.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.58 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 1819565 120 - 6413771 AAGCCCGUGACGAGGAUUGAACUCGUGACCUCACCCUUACCAAGGGUGUGCUCUACCACUGAGCCACACGGGCGAAAUUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAU ..((((((((((((.......))))).....((((((.....)))))).((((.......))))..)))))))....................((((((((.((.....)).)))))))) ( -38.80) >anava.0 6278914 120 - 6365727 AAGCCCGUGACGAGGAUUGAACUCGUGACCUCACCCUUACCAAGGGUGUGCUCUACCACUGAGCCACACGGGCGGAAGUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAU ..((((((((((((.......))))).....((((((.....)))))).((((.......))))..)))))))....................((((((((.((.....)).)))))))) ( -38.80) >consensus AAGCCCGUGACGAGGAUUGAACUCGUGACCUCACCCUUACCAAGGGUGUGCUCUACCACUGAGCCACACGGGCGAAAGUUAUUUAAAUCUUAGAUUUUGAACUAGGUUAUAAUUCAAAAU ..((((((((((((.......))))).....((((((.....)))))).((((.......))))..)))))))....................((((((((.((.....)).)))))))) (-38.80 = -38.80 + 0.00)

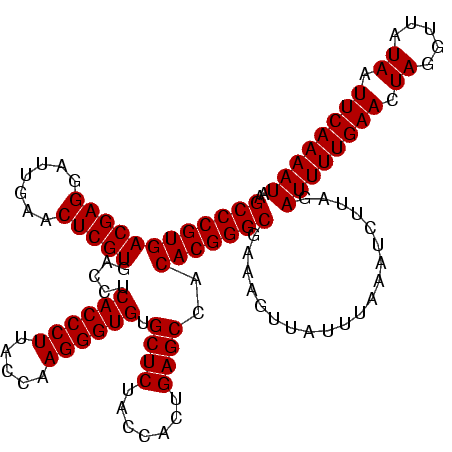
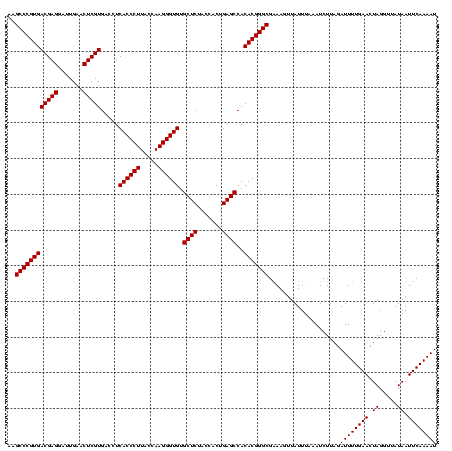
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Wed Apr 5 20:05:50 2006