
| Sequence ID | nosto.0 |
|---|---|
| Location | 1,592,156 – 1,592,223 |
| Length | 67 |
| Max. P | 0.966859 |

| Location | 1,592,156 – 1,592,223 |
|---|---|
| Length | 67 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 73.55 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -18.95 |
| Energy contribution | -16.18 |
| Covariance contribution | -2.77 |
| Combinations/Pair | 1.79 |
| Mean z-score | -0.58 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
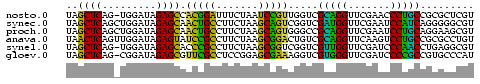
>nosto.0 1592156 67 + 6413771 UAGCUCAG-UGGAUAGAGCCACGGAUUUCUAAUCCGUUGGUCGCAGGUUCGAACCCUGCCGCGCUCGU ........-......((((.((((((.....))))))..((.(((((.......))))).)))))).. ( -21.40) >synec.0 1813573 68 + 2434428 UAGCUCAGCUGGAUAGAGCAACUGCCUUCUAAGCAGUCGGUCGAUGGUUCGAAUCCAUCAGGGGGCGU ..((((..((.....((.(.(((((.......))))).).))(((((.......))))))).)))).. ( -21.70) >proch.0 1203672 68 + 1709204 UAGCUCAGCUGGAUAGAGCAACUGCCUUCUAAGCAGUGGGCCGCAGGUUCGAAUCCUGCAGGAAGCGU ..((((.(((......))).(((((.......))))))))).(((((.......)))))......... ( -21.70) >anava.0 24136 68 - 6365727 UAACUCAGUUGGAUAGAGUAUCCGCCUUCUAAGCGGACUGUCGCAGGUUCAAGUCCUGCCGCGCCUGU .....(((.(((((((....(((((.......))))))))))(((((.......)))))..)).))). ( -20.80) >synel.0 722816 67 - 2696255 UAGCUCAG-UGGAUAGAGCACCCGCCUUCUAAGCGGUCGGUCGUUGGUUCGAUCCCAACCUGAGGCGU ...(((((-......((.(..((((.......))))..).))(((((.......)))))))))).... ( -20.20) >gloev.0 3392744 67 + 4659019 UAGCUCAG-CGGAUAGAGCGUUCGCCUCCGGAGCGAAAGGUCGUGGGUUCGAUCCCCGCCGUGCCCAU ..((((..-......))))..........((.(((...((.((.(((......))))))).))))).. ( -21.60) >consensus UAGCUCAG_UGGAUAGAGCAACCGCCUUCUAAGCGGUCGGUCGCAGGUUCGAACCCUGCCGGGCGCGU ..((((.........)))).(((((.......))))).....(((((.......)))))......... (-18.95 = -16.18 + -2.77)
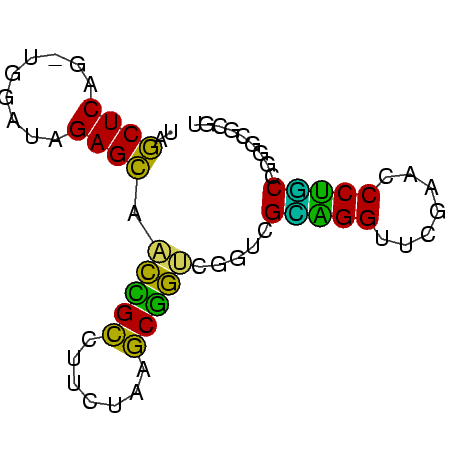
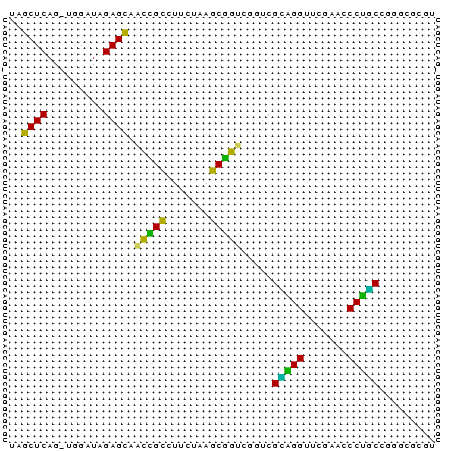
| Location | 1,592,156 – 1,592,223 |
|---|---|
| Length | 67 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 73.55 |
| Mean single sequence MFE | -22.33 |
| Consensus MFE | -20.71 |
| Energy contribution | -18.13 |
| Covariance contribution | -2.58 |
| Combinations/Pair | 1.73 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
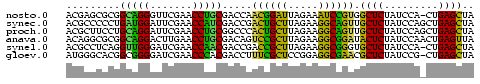
>nosto.0 1592156 67 - 6413771 ACGAGCGCGGCAGGGUUCGAACCUGCGACCAACGGAUUAGAAAUCCGUGGCUCUAUCCA-CUGAGCUA .........(((((.......))))).....((((((.....))))))(((((......-..))))). ( -22.50) >synec.0 1813573 68 - 2434428 ACGCCCCCUGAUGGAUUCGAACCAUCGACCGACUGCUUAGAAGGCAGUUGCUCUAUCCAGCUGAGCUA ..(((..((((((((.((((....)))).((((((((.....)))))))).))))).)))..).)).. ( -20.60) >proch.0 1203672 68 - 1709204 ACGCUUCCUGCAGGAUUCGAACCUGCGGCCCACUGCUUAGAAGGCAGUUGCUCUAUCCAGCUGAGCUA .......(((((((.......)))))))...((((((.....)))))).((((.........)))).. ( -21.80) >anava.0 24136 68 + 6365727 ACAGGCGCGGCAGGACUUGAACCUGCGACAGUCCGCUUAGAAGGCGGAUACUCUAUCCAACUGAGUUA .(((.....(((((.......)))))((..(((((((.....)))))))..)).......)))..... ( -21.60) >synel.0 722816 67 + 2696255 ACGCCUCAGGUUGGGAUCGAACCAACGACCGACCGCUUAGAAGGCGGGUGCUCUAUCCA-CUGAGCUA ..((.(((((((((.......)))))((.((.(((((.....))))).)).))......-)))))).. ( -23.00) >gloev.0 3392744 67 - 4659019 AUGGGCACGGCGGGGAUCGAACCCACGACCUUUCGCUCCGGAGGCGAACGCUCUAUCCG-CUGAGCUA ...(((.((((((((.(((......))))).((((((.....))))))........)))-))).))). ( -24.50) >consensus ACGCGCCCGGCAGGAUUCGAACCUGCGACCGACCGCUUAGAAGGCGGUUGCUCUAUCCA_CUGAGCUA .........(((((.......))))).....((((((.....)))))).((((.........)))).. (-20.71 = -18.13 + -2.58)
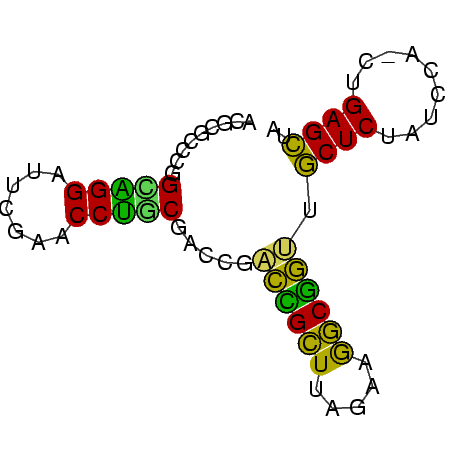
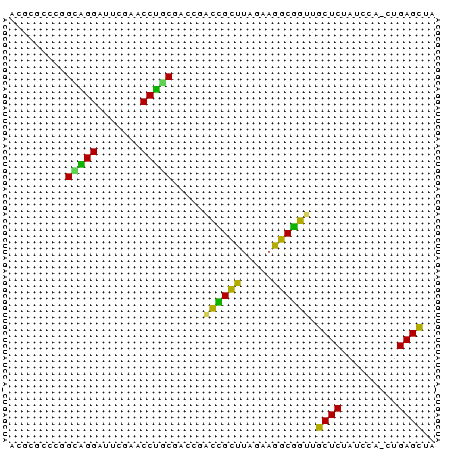
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Wed Apr 5 20:05:42 2006