
| Sequence ID | nosto.0 |
|---|---|
| Location | 610,396 – 610,474 |
| Length | 78 |
| Max. P | 0.999900 |

| Location | 610,396 – 610,474 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -32.49 |
| Energy contribution | -30.77 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.53 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
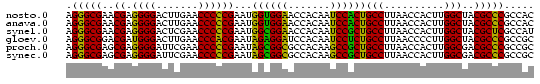
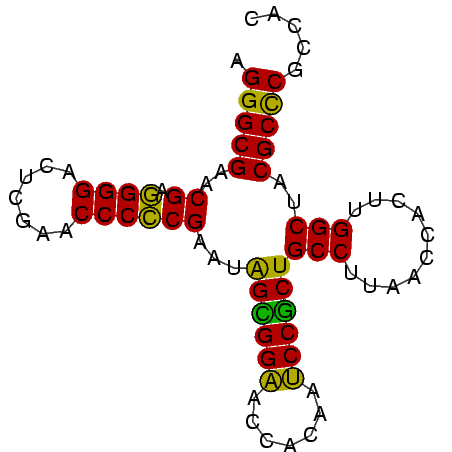
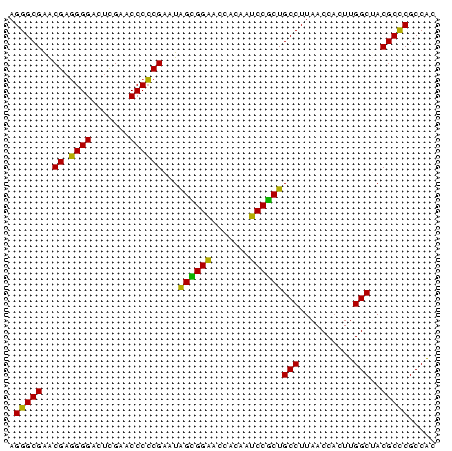
>nosto.0 610396 78 + 6413771 AGGGCGAACGAGGGGACUUGAACCCCCGAAUGGUGGAACCACAAUCCACUGCCUUAACCACUUGGCUACGCCCGCCAC .(((((..((.((((.......))))))...((((((.......))))))(((..........)))..)))))..... ( -29.80) >anava.0 3626038 78 + 6365727 AGGGCGAACGAGGGGACUUGAACCCCCGAAUGGUGGAACCACAAUCCACUGCCUUAACCACUUGGCUACGCCCGCCAC .(((((..((.((((.......))))))...((((((.......))))))(((..........)))..)))))..... ( -29.80) >synel.0 1883459 78 + 2696255 AGGGCGAACGAGGGGACUCGAACCCCCGAAUGGCGGAACCACAAUCCGCUGCCUUAACCACUUGGCUACGCUCGCCAU ..(((((.((.((((........))))....((((((.......))))))(((..........)))..)).))))).. ( -30.60) >gloev.0 686428 78 - 4659019 AGGGCGGACGAUGGGACUUGAACCCACGAAUAGAGGAUCCACAAUCCUCUGCCUUAACCCCUUGGCUACGCCCGCCGC .(((((..((.((((.......))))))...(((((((.....)))))))(((..........)))..)))))..... ( -29.20) >proch.0 391505 78 - 1709204 AGGGCGAGCGAGGGGAUUCGAACCCCCGAAUAGCGGCGCCACAAGCCGCUGCCUUAACCACUUGGCGACGCCCGCCGC ..((((.(((.((((........))))....((((((.......))))))(((..........)))..))).)))).. ( -35.80) >synec.0 856671 78 - 2434428 AGGGCGAGCGAGGGGAUUCGAACCCCCGAAUAGCGGCGCCACAAGCCGCUGCCUUAACCACUUGGCGACGCCCGCCGC ..((((.(((.((((........))))....((((((.......))))))(((..........)))..))).)))).. ( -35.80) >consensus AGGGCGAACGAGGGGACUCGAACCCCCGAAUAGCGGAACCACAAUCCGCUGCCUUAACCACUUGGCUACGCCCGCCAC .(((((..((.((((.......))))))...((((((.......))))))(((..........)))..)))))..... (-32.49 = -30.77 + -1.72)
| Location | 610,396 – 610,474 |
|---|---|
| Length | 78 |
| Sequences | 6 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -34.24 |
| Energy contribution | -32.42 |
| Covariance contribution | -1.83 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
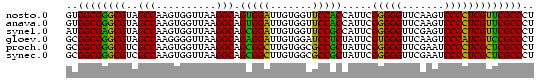
>nosto.0 610396 78 + 6413771 GUGGCGGGCGUAGCCAAGUGGUUAAGGCAGUGGAUUGUGGUUCCACCAUUCGGGGGUUCAAGUCCCCUCGUUCGCCCU ..(((((((((((((....))))).....(((((.......))))).....(((((......))))).)))))))).. ( -31.00) >anava.0 3626038 78 + 6365727 GUGGCGGGCGUAGCCAAGUGGUUAAGGCAGUGGAUUGUGGUUCCACCAUUCGGGGGUUCAAGUCCCCUCGUUCGCCCU ..(((((((((((((....))))).....(((((.......))))).....(((((......))))).)))))))).. ( -31.00) >synel.0 1883459 78 + 2696255 AUGGCGAGCGUAGCCAAGUGGUUAAGGCAGCGGAUUGUGGUUCCGCCAUUCGGGGGUUCGAGUCCCCUCGUUCGCCCU ..(((((((((((((....))))).....(((((.......))))).....(((((......))))).)))))))).. ( -33.50) >gloev.0 686428 78 - 4659019 GCGGCGGGCGUAGCCAAGGGGUUAAGGCAGAGGAUUGUGGAUCCUCUAUUCGUGGGUUCAAGUCCCAUCGUCCGCCCU ..(((((((((((((....)))))....((((((((...))))))))....(((((.......))))))))))))).. ( -34.20) >proch.0 391505 78 - 1709204 GCGGCGGGCGUCGCCAAGUGGUUAAGGCAGCGGCUUGUGGCGCCGCUAUUCGGGGGUUCGAAUCCCCUCGCUCGCCCU ((((((((((((((.((((.(((.....))).)))))))))))).......((((((....)))))).))).)))... ( -39.00) >synec.0 856671 78 - 2434428 GCGGCGGGCGUCGCCAAGUGGUUAAGGCAGCGGCUUGUGGCGCCGCUAUUCGGGGGUUCGAAUCCCCUCGCUCGCCCU ((((((((((((((.((((.(((.....))).)))))))))))).......((((((....)))))).))).)))... ( -39.00) >consensus GCGGCGGGCGUAGCCAAGUGGUUAAGGCAGCGGAUUGUGGUUCCGCCAUUCGGGGGUUCAAGUCCCCUCGUUCGCCCU ..((((((((..(((..........))).(((((.......))))).....(((((.......))))))))))))).. (-34.24 = -32.42 + -1.83)
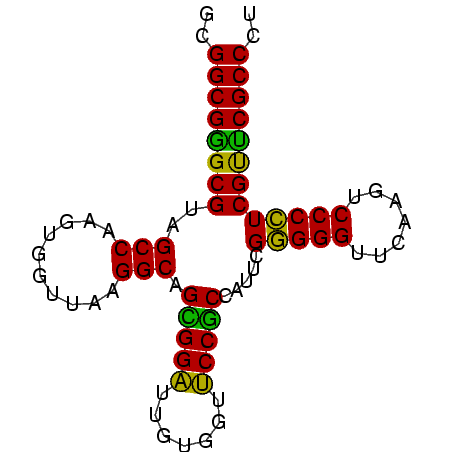
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Wed Apr 19 19:58:21 2006