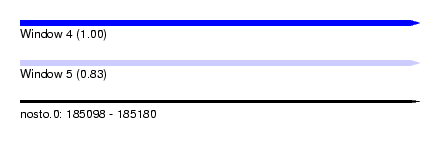
| Sequence ID | nosto.0 |
|---|---|
| Location | 185,098 – 185,180 |
| Length | 82 |
| Max. P | 0.997670 |

| Location | 185,098 – 185,180 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 84 |
| Reading direction | forward |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -34.60 |
| Energy contribution | -32.35 |
| Covariance contribution | -2.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
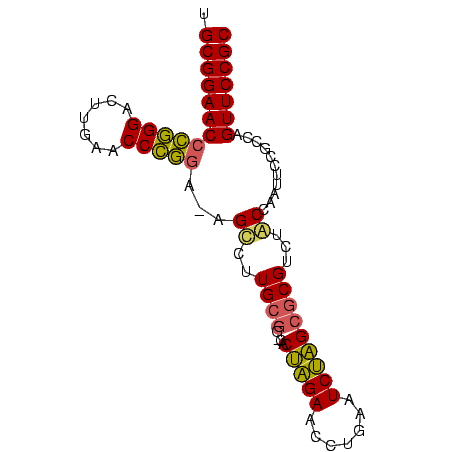
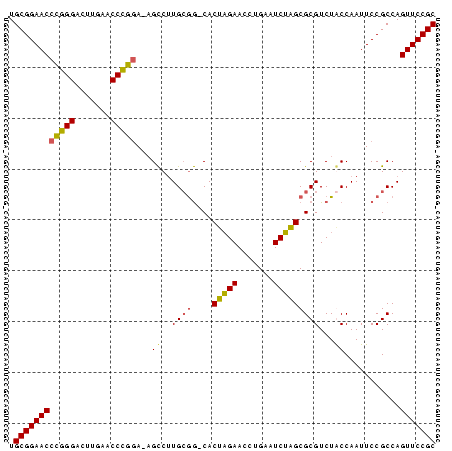
>nosto.0 185098 82 + 6413771 GCGGAACUGGCGGAAUUGGCAGACGCGCUAGAUUCAGGUUCUAGUG-CCGCAAGGCU-UCCGGGUUCAAGUCCCGGGUUCCGCA ((((((((.(.(((.(((...(((.(((((((.......))))))(-((....))).-...).)))))).)))).)))))))). ( -38.40) >anava.0 1777232 82 + 6365727 GCGGAACUGGCGGAAUUGGCAGACGCGCUAGAUUCAGGUUCUAGUG-CCGCAAGGCU-UCCGGGUUCAAGUCCCGGGUUCCGCA ((((((((.(.(((.(((...(((.(((((((.......))))))(-((....))).-...).)))))).)))).)))))))). ( -38.40) >synel.0 911177 84 + 2696255 GCGGAACUGGCGGAAUUGGUAGACGCGCUAGAUUCAGGUUCUAGUGGUUUCACGACUGUCCGGGUUCAAGUCCCGGGUUCCGCA (((((((((((((..(((..((((.(((((((.......)))))))))))..))))))))((((.......)))))))))))). ( -38.10) >gloev.0 3717168 82 + 4659019 GCGGAACUGGCGGAAUUGGUAGACGCGCAUGAUUCAGGUUCAUGUG-CCGCAAGGCG-UGGAGGUUCAAGUCCUCUGUUCCGCA (((((((.((.(((.((((....(((((((((.......))))))(-((....))))-)).....)))).))).))))))))). ( -37.90) >consensus GCGGAACUGGCGGAAUUGGCAGACGCGCUAGAUUCAGGUUCUAGUG_CCGCAAGGCU_UCCGGGUUCAAGUCCCGGGUUCCGCA (((((((..((((.....((....))((((((.......))))))..))))........(((((.......)))))))))))). (-34.60 = -32.35 + -2.25)

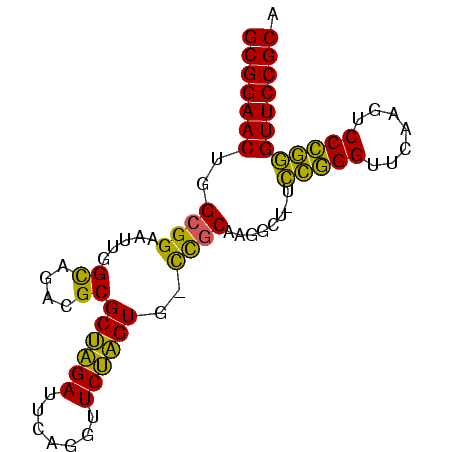
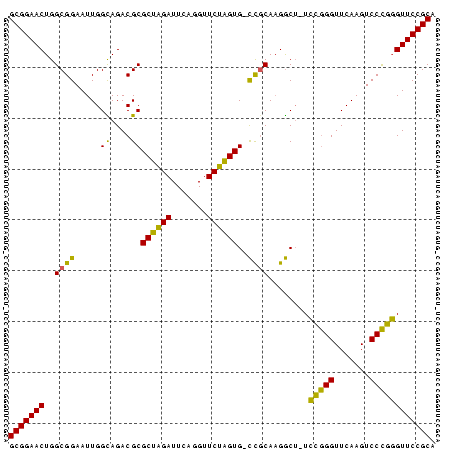
| Location | 185,098 – 185,180 |
|---|---|
| Length | 82 |
| Sequences | 4 |
| Columns | 84 |
| Reading direction | reverse |
| Mean pairwise identity | 88.35 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -23.95 |
| Energy contribution | -23.20 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>nosto.0 185098 82 + 6413771 UGCGGAACCCGGGACUUGAACCCGGA-AGCCUUGCGG-CACUAGAACCUGAAUCUAGCGCGUCUGCCAAUUCCGCCAGUUCCGC .((((((((((((.......))))).-........((-(.(((((.......))))).((....)).......))).))))))) ( -29.20) >anava.0 1777232 82 + 6365727 UGCGGAACCCGGGACUUGAACCCGGA-AGCCUUGCGG-CACUAGAACCUGAAUCUAGCGCGUCUGCCAAUUCCGCCAGUUCCGC .((((((((((((.......))))).-........((-(.(((((.......))))).((....)).......))).))))))) ( -29.20) >synel.0 911177 84 + 2696255 UGCGGAACCCGGGACUUGAACCCGGACAGUCGUGAAACCACUAGAACCUGAAUCUAGCGCGUCUACCAAUUCCGCCAGUUCCGC .((((((((((((.......)))))..((.((((......(((((.......))))))))).)).............))))))) ( -27.40) >gloev.0 3717168 82 + 4659019 UGCGGAACAGAGGACUUGAACCUCCA-CGCCUUGCGG-CACAUGAACCUGAAUCAUGCGCGUCUACCAAUUCCGCCAGUUCCGC .(((((((...(((.(((........-(((...)))(-(.(((((.......))))).))......))).)))....))))))) ( -26.30) >consensus UGCGGAACCCGGGACUUGAACCCGGA_AGCCUUGCGG_CACUAGAACCUGAAUCUAGCGCGUCUACCAAUUCCGCCAGUUCCGC .((((((((((((.......)))))...((..((((....(((((.......)))))))))...))...........))))))) (-23.95 = -23.20 + -0.75)
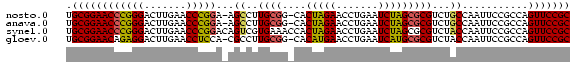
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Wed Apr 19 19:58:18 2006