
| Sequence ID | nosto.0 |
|---|---|
| Location | 56,433 – 56,519 |
| Length | 86 |
| Max. P | 0.999530 |

| Location | 56,433 – 56,519 |
|---|---|
| Length | 86 |
| RNAz 1.0pre Sequences | 4 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -28.64 |
| Energy contribution | -28.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -4.16 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
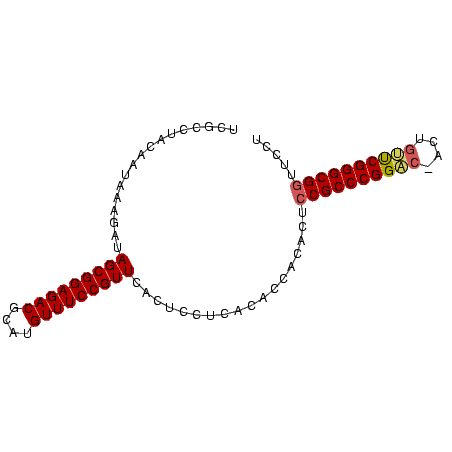
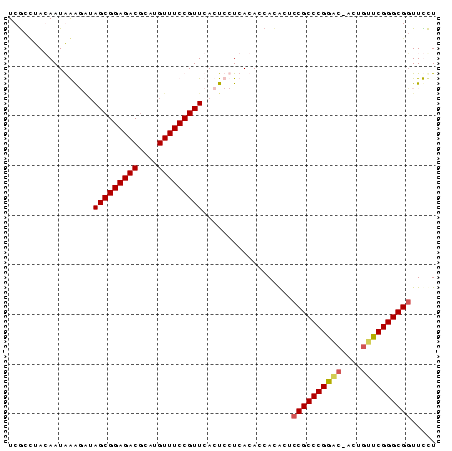
>nosto.0 56433 86 + 6413771 UCGCCUACAAUAGAUAUAGCGGAGACGCAUGUUUCCGUUCACUCCUCACACCACACUCCGCCCGGACUACUGUUCGGGCGGUUCCU ............((...(((((((((....)))))))))...)).............((((((((((....))))))))))..... ( -31.00) >anava.0 3283406 86 + 6365727 UCGCCUACAAUAGAUAUAGCGGAGACGCAUGUUUCCGUUCACUCCUCACACCACACUCCGCCCGGACUACUGUUCGGGCGGUUCCU ............((...(((((((((....)))))))))...)).............((((((((((....))))))))))..... ( -31.00) >gloev.0 3727265 83 - 4659019 UCGCAUAGAAUGAAGGAAGCGGAGACGCAUGUUUCCGUUCACUCCUCACACCAUGUUCCGCCCGGUC---GCUCCGGGCGGUUUCU ..((((.(..(((.((((((((((((....)))))))))...))))))..).)))).((((((((..---...))))))))..... ( -35.70) >synel.0 2417281 85 + 2696255 UCACUUAGAAUGAGGUUAGCGGAGACAAAUGUUUCCGUUCACUCCUCACACCACACUUCGCCCGGGC-AACGCUCGGGCGUUUUUU ..........(((((..(((((((((....)))))))))....)))))..........(((((((((-...)))))))))...... ( -35.50) >consensus UCGCCUACAAUAAAGAUAGCGGAGACGCAUGUUUCCGUUCACUCCUCACACCACACUCCGCCCGGAC_ACUGUUCGGGCGGUUCCU .................(((((((((....)))))))))..................((((((((((....))))))))))..... (-28.64 = -28.95 + 0.31)

| Location | 56,433 – 56,519 |
|---|---|
| Length | 86 |
| Sequences | 4 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 81.75 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -25.26 |
| Energy contribution | -26.70 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
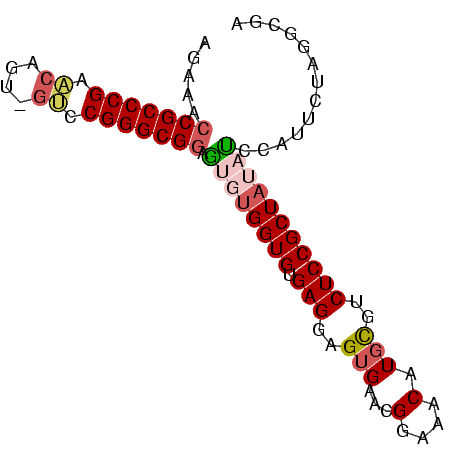
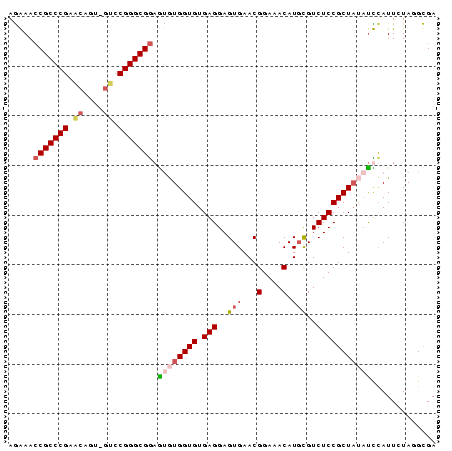
>nosto.0 56433 86 + 6413771 AGGAACCGCCCGAACAGUAGUCCGGGCGGAGUGUGGUGUGAGGAGUGAACGGAAACAUGCGUCUCCGCUAUAUCUAUUGUAGGCGA ......((((...(((((((....(((((((((((.(((.....(....)....))))))).)))))))....))))))).)))). ( -31.90) >anava.0 3283406 86 + 6365727 AGGAACCGCCCGAACAGUAGUCCGGGCGGAGUGUGGUGUGAGGAGUGAACGGAAACAUGCGUCUCCGCUAUAUCUAUUGUAGGCGA ......((((...(((((((....(((((((((((.(((.....(....)....))))))).)))))))....))))))).)))). ( -31.90) >gloev.0 3727265 83 - 4659019 AGAAACCGCCCGGAGC---GACCGGGCGGAACAUGGUGUGAGGAGUGAACGGAAACAUGCGUCUCCGCUUCCUUCAUUCUAUGCGA .....((((((((...---..))))))))..(((((.((((((...(((((((.((....)).)))).))))))))).)))))... ( -36.20) >synel.0 2417281 85 + 2696255 AAAAAACGCCCGAGCGUU-GCCCGGGCGAAGUGUGGUGUGAGGAGUGAACGGAAACAUUUGUCUCCGCUAACCUCAUUCUAAGUGA ......((((((.((...-)).)))))).((((.(((....((((((((.(....).)))).))))....))).))))........ ( -29.90) >consensus AGAAACCGCCCGAACAGU_GUCCGGGCGGAGUGUGGUGUGAGGAGUGAACGGAAACAUGCGUCUCCGCUAUAUCCAUUCUAGGCGA .....(((((((.((....)).))))))).((((((((.(((..(((...(....).)))..)))))))))))............. (-25.26 = -26.70 + 1.44)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Wed Apr 19 19:58:17 2006