 Pscan
Pscan
 DAS
DAS
 Department
Department
 Miklos
Miklos

"DAS" - Transmembrane Prediction server
Computation is in progress.
It takes a minute or two typically.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
Done
Your query is:
MSAVTESVLESIISPVTMSEFLEEYWPVKPLVARGEVERFTSIPGFEKVRTLENVLAIYN
NPVMVVGDAVIEESEGITDRFLVSPAEALEWYEKGAALEFDFTDLFIPQVRRWIEKLKAE
LRLPAGTSSKAIVYAAKNGGGFKAHFDAYTNLIFQIQGEKTWKLAKNENVSNPMQHYDLS
EAPYYPDDLQSYWKGDPPKEDLPDAEIVNLTPGTMLYLPRGLWHSTKSDQATLALNITFG
QPAWLDLMLAALRKKLISDNRFRELAVNHQSLHESSKSELNGYLESLIQTLSENAETLTP
EQIFQSQDSDFDPYQSTQLVFRQLLTSYKF
Potential transmembrane segments
Start Stop Length ~ Cutoff
11 13 3 ~ 1.7
63 64 2 ~ 1.7
248 250 3 ~ 1.7
The DAS curve for your query:
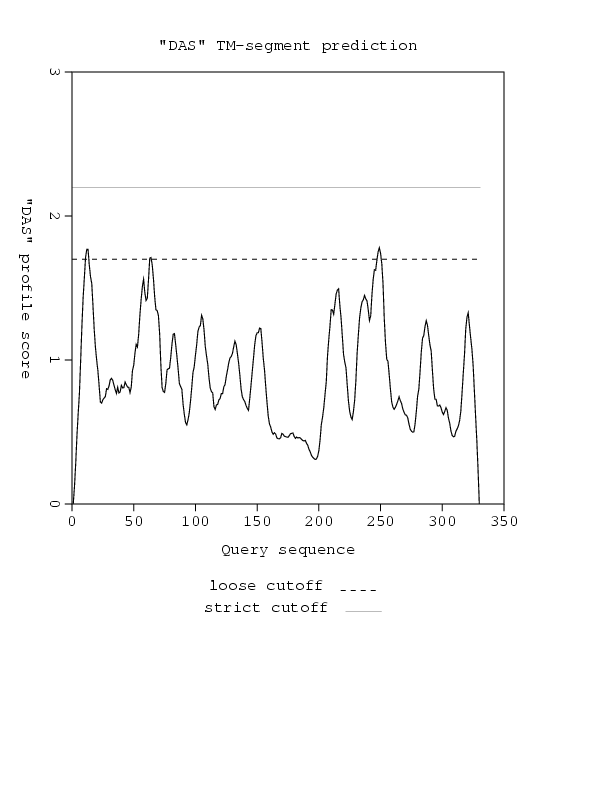
The DAS curve of the query is also available in
PostScript
format.
You may wish to compare your curve with those obtained for the
test set. It gives you an idea about the
confidence of the prediction.
"DAS" curves of the test set
These curves are obtained by pairwise comparison of the proteins in the
test set in "each against the rest" fashion. There are two cutoffs
indicated on the plots: a "strict" one at 2.2 DAS score, and a "loose" one
at 1.7. The hit at 2.2 is informative in terms of the number of matching
segments, while a hit at 1.7 gives the actual location of the
transmembrane segment. The segments reported in the "FT" records of the
SwissProt database are marked at 1.0 DAS score ("FT lines").
ALKB_PSEOL,
ATPL_ECOLI,
COX2_PARDE,
COX3_PARDE,
CX1B_PARDE,
CYDA_ECOLI,
CYDB_ECOLI,
CYOA_ECOLI,
CYOB_ECOLI,
CYOC_ECOLI,
CYOD_ECOLI,
CYOE_ECOLI,
DHG_ECOLI,
DMSC_ECOLI,
DSBB_ECOLI,
ENVZ_ECOLI,
EXBB_ECOLI,
EXBD_ECOLI,
FTSH_ECOLI,
FTSL_ECOLI,
FUCP_ECOLI,
GLPT_ECOLI,
HISM_SALTY,
HISQ_SALTY,
HOXN_ALCEU,
IMMA_CITFR,
KDPD_ECOLI,
KGTP_ECOLI,
LACY_ECOLI,
LSPA_ECOLI,
MALG_ECOLI,
MELB_ECOLI,
MOTA_ECOLI,
MOTB_ECOLI,
MTR_ECOLI,
OPPB_SALTY,
OPPC_SALTY,
PHOR_ECOLI,
RHAT_ECOLI,
SECD_ECOLI,
SECE_ECOLI,
SECY_ECOLI,
TOLQ_ECOLI,
TOLR_ECOLI

