| View Alignment |
Fold Library |
Template Length |
Model
| PSSM E_value |
SAWTED E-value |
Biotext |
Class |
Fold |
SuperFamily |
Family |
Protein |
Species |
ID |
Region |
CS |
PSSM Type
|
 |
c1pj5a_ 14%i.d. |
827 |
|
1.31e-09 |
1 | n/a |
not in SCOP 1.53 |
PDB header: oxidoreductase. |
Chain: A: PDB Molecule:n,n-dimethylglycine oxidase;. |
. |
PDBTitle: crystal structure of dimethylglycine oxidase of2 arthrobacter globiformis in complex with acetate
.
| - |
1pj5 |
a
|
0.000 |
R1D |
 |
c1ko3a_ 15%i.d. |
230 |

|
0.0473 |
1 | n/a |
Alpha and beta proteins (a+b) |
Metallo-hydrolase |
Metallo-hydrolase |
homol. to d1a7ta_ |
- |
- |
1ko3 |
a
|
0.000 |
R1D |
 |
d1egaa1 11%i.d. |
179 |
|
0.614 |
1 | 0.92

|
Alpha and beta proteins (a/b) |
P-loop containing nucleotide triphosphate hydrolases |
P-loop containing nucleotide triphosphate hydrolases |
G proteins |
GTPase Era, N-terminal domain |
Escherichia coli |
1ega |
a:4-182
|
0.500 |
R1D |
 |
c1ky3a_ 15%i.d. |
162 |
|
2.35 |
1 | 1.00
|
Alpha and beta proteins (a/b) |
P-loop containing nucleotide triphosphate hydrolases |
P-loop containing nucleotide triphosphate hydrolases |
homol. to d3raba_ |
- |
- |
1ky3 |
a
|
0.000 |
R1D |
 |
c1egaa_ 10%i.d. |
292 |
|
2.54 |
1 | 0.92

|
not in SCOP 1.53 |
PDB header: hydrolase. |
Chain: A: PDB Molecule:gtp-binding protein era;. |
. |
PDBTitle: crystal structure of a widely conserved gtpase era
.
| - |
1ega |
a
|
0.000 |
R1D |
 |
c1ddka_ 9%i.d. |
220 |
|
2.91 |
1 | 1.00
|
Alpha and beta proteins (a+b) |
Metallo-hydrolase |
Metallo-hydrolase |
homol. to d1a7ta_ |
- |
- |
1ddk |
a
|
0.000 |
R1D |
 |
c1gs5a_ 14%i.d. |
258 |
|
2.95 |
0.945

| 0.74

|
not in SCOP 1.53 |
PDB header: kinase. |
Chain: A: PDB Molecule:acetylglutamate kinase;. |
. |
PDBTitle: n-acetyl-l-glutamate kinase
from escherichia coli complexed2 with its substrate n-acetylglutamate
and its substrate3 analog amppnp . | - |
1gs5 |
a
|
0.000 |
R1D |
 |
d1gdoa_ 17%i.d. |
238 |
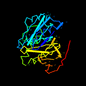
|
3.69 |
0.68

| 0.63

|
Alpha and beta proteins (a+b) |
N-terminal nucleophile aminohydrolases (Ntn hydrolases) |
N-terminal nucleophile aminohydrolases (Ntn hydrolases) |
Class II glutamine amidotransferases |
Glucosamine 6-phosphate synthase, N-terminal domain |
Escherichia coli |
1gdo |
a:
|
0.500 |
R1D |
 |
d1cm0a_ 12%i.d. |
162 |

|
4.51 |
1 | n/a |
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Histone acetyltransferase domain of P300/CBP associating factor, PCAF |
Human (Homo sapiens) |
1cm0 |
a:
|
0.806 |
R1D |
 |
d1mfoa_ 11%i.d. |
262 |
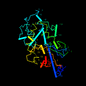
|
5.62 |
1 | 1.00
|
Multi-domain proteins (alpha and beta) |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase, class A |
Mycobacterium fortuitum |
1mfo |
a:
|
0.723 |
R1D |
 |
d1bza__ 14%i.d. |
257 |
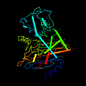
|
5.81 |
1 | 1.00
|
Multi-domain proteins (alpha and beta) |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase, class A |
Escherichia coli, TOHO-1 |
1bza |
-
|
0.721 |
1D |
 |
c1h3da_ 14%i.d. |
288 |

|
5.96 |
0.951

| n/a |
not in SCOP 1.53 |
PDB header: transferase. |
Chain: A: PDB Molecule:atp-phosphoribosyltransferase;. |
. |
PDBTitle: structure of the e.coli atp-phosphoribosyltransferase
.
| - |
1h3d |
a
|
0.000 |
R1D |
 |
d1mrg__ 10%i.d. |
246 |
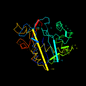
|
6.17 |
1 | 1.00
|
Alpha and beta proteins (a+b) |
Ribosome inactivating proteins (RIP) |
Ribosome inactivating proteins (RIP) |
Plant cytotoxins |
alpha-Momorcharin (momordin) |
Bitter gourd (Momordica charantia) |
1mrg |
-
|
0.790 |
R1D |
 |
d1buea_ 11%i.d. |
265 |
|
6.29 |
1 | 1.00
|
Multi-domain proteins (alpha and beta) |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase, class A |
Enterobacter cloacae, NMC-A carbapenemase |
1bue |
a:
|
0.947 |
1D |
 |
d4blma_ 15%i.d. |
256 |
|
7.75 |
0.883

| 1.00
|
Multi-domain proteins (alpha and beta) |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase/D-ala carboxypeptidase |
beta-Lactamase, class A |
Bacillus licheniformis |
4blm |
a:
|
0.778 |
1D |
 |
c1iq6a_ 13%i.d. |
132 |

|
8.23 |
1 | n/a |
not in SCOP 1.53 |
PDB header: lyase. |
Chain: A: PDB Molecule:(r)-specific enoyl-coa hydratase;. |
. |
PDBTitle: (r)-hydratase from a. caviae involved in pha biosynthesis
.
| - |
1iq6 |
a
|
0.000 |
R1D |
 |
c1fmta_ 13%i.d. |
308 |
|
9.09 |
0.917

| 0.77

|
not in SCOP 1.53 |
PDB header: formyltransferase. |
Chain: A: PDB Molecule:methionyl-trna fmet formyltransferase;. |
. |
PDBTitle: methionyl-trnafmet formyltransferase from escherichia coli
.
| - |
1fmt |
a
|
0.000 |
R1D |
 |
c1m61a_ 14%i.d. |
259 |
|
9.23 |
0.246

| n/a |
Alpha and beta proteins (a+b) |
SH2-like |
SH2 domain |
homol. to d1a81a_ |
- |
- |
1m61 |
a
|
0.000 |
R1D |
 |
d1fmta2 12%i.d. |
200 |
|
11.6 |
0.917

| 0.77

|
Alpha and beta proteins (a/b) |
Formyltransferase |
Formyltransferase |
Formyltransferase |
Methionyl-tRNAfmet formyltransferase |
Escherichia coli |
1fmt |
a:1-206
|
0.000 |
R1D |
 |
c1eeja_ 13%i.d. |
216 |
|
12.5 |
1 | 1.00
|
not in SCOP 1.53 |
PDB header: isomerase. |
Chain: A: PDB Molecule:thiol:disulfide interchange protein;. |
. |
PDBTitle: crystal structure of the protein disulfide bond isomerase,2 dsbc, from escherichia coli
.
| - |
1eej |
a
|
0.000 |
R1D |