| View Alignment |
Fold Library |
Template Length |
Model
| PSSM E_value |
SAWTED E-value |
Biotext |
Class |
Fold |
SuperFamily |
Family |
Protein |
Species |
ID |
Region |
CS |
PSSM Type
|
 |
c1b9ka_ 16%i.d. |
237 |
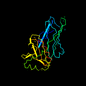
|
0.123 |
1 | 1.00
|
not in SCOP 1.53 |
PDB header: endocytosis/exocytosis. |
Chain: A: PDB Molecule:alpha-adaptin appendage domain;. |
. |
PDBTitle: alpha-adaptin appendage domain, from clathrin adaptor ap2
.
| - |
1b9k |
a
|
0.000 |
1D |
 |
d1pdkb_ 18%i.d. |
149 |
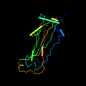
|
0.729 |
0.759

| 0.74

|
All beta proteins |
Common fold of diphtheria toxin/transcription factors/cytochrome f |
Bacterial adhesins |
Pilus subunits |
PapK pilus subunit |
Escherichia coli |
1pdk |
b:
|
0.901 |
R1D |
 |
d6prch1 19%i.d. |
222 |
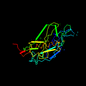
|
1.08 |
0.961

| 0.83

|
All beta proteins |
Photosynthetic reaction centre, H-chain, cytoplasmic domain |
Photosynthetic reaction centre, H-chain, cytoplasmic domain |
Photosynthetic reaction centre, H-chain, cytoplasmic domain |
Photosynthetic reaction centre |
Rhodopseudomonas viridis |
6prc |
h:37-258
|
0.000 |
R1D |
 |
d1i16__ 15%i.d. |
130 |
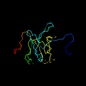
|
2.53 |
1 | 1.00
|
All beta proteins |
PDZ domain-like |
PDZ domain-like |
Interleukin 16 |
Interleukin 16 |
Human (Homo sapiens) |
1i16 |
-
|
1.000 |
R1D |
 |
d1wba__ 16%i.d. |
171 |
|
3.03 |
1 | 0.85

|
All beta proteins |
beta-Trefoil |
STI-like |
Kunitz (STI) inhibitors |
Winged bean albumin 1 |
Goa bean (Psophocarpus tetragonolobus) |
1wba |
-
|
0.569 |
R1D |
 |
d1obwa_ 22%i.d. |
175 |
|
3.05 |
0.863

| 1.00
|
All beta proteins |
OB-fold |
Inorganic pyrophosphatase |
Inorganic pyrophosphatase |
Inorganic pyrophosphatase |
Escherichia coli |
1obw |
a:
|
0.702 |
R1D |
 |
c1e42a_ 13%i.d. |
233 |
|
3.42 |
1 | n/a |
not in SCOP 1.53 |
PDB header: endocytosis. |
Chain: A: PDB Molecule:beta2-adaptin;. |
. |
PDBTitle: beta2-adaptin appendage domain, from clathrin adaptor ap2
.
| - |
1e42 |
a
|
0.000 |
1D |
 |
d1wioa2 18%i.d. |
113 |

|
3.42 |
0.857

| 0.76

|
All beta proteins |
Immunoglobulin-like beta-sandwich |
Immunoglobulin |
V set domains (antibody variable domain-like) |
CD4 |
Human (Homo sapiens) |
1wio |
a:179-291
|
0.423 |
3D |
 |
d1kacb_ 10%i.d. |
124 |

|
3.8 |
0.635

| 0.70

|
All beta proteins |
Immunoglobulin-like beta-sandwich |
Immunoglobulin |
V set domains (antibody variable domain-like) |
Coxsackie virus and adenovirus receptor (Car), domain 1 |
Human (Homo sapiens) |
1kac |
b:
|
0.400 |
3D |
 |
d1avwb_ 15%i.d. |
171 |
|
4.26 |
0.946

| 0.85

|
All beta proteins |
beta-Trefoil |
STI-like |
Kunitz (STI) inhibitors |
Soybean trypsin inhibitor |
Soybean (Glycine max) |
1avw |
b:
|
0.652 |
1D |
 |
d1b9ma2 16%i.d. |
135 |

|
4.9 |
0.896

| 1.00
|
All beta proteins |
OB-fold |
C-terminal domain of molybdate-dependent transcriptional regulator ModE |
C-terminal domain of molybdate-dependent transcriptional regulator ModE |
C-terminal domain of molybdate-dependent transcriptional regulator ModE |
Escherichia coli |
1b9m |
a:127-262
|
0.500 |
1D |
 |
d1a8l_2 15%i.d. |
107 |

|
5.65 |
1 | 0.82

|
Alpha and beta proteins (a/b) |
Thioredoxin fold |
Thioredoxin-like |
Protein disulfide isomerase |
Protein disulfide isomerase |
Pyrococcus furiosus |
1a8l |
120-226
|
0.500 |
R1D |
 |
d1pama2 18%i.d. |
104 |

|
5.93 |
0.867

| 0.83

|
All beta proteins |
Prealbumin-like |
Starch-binding domain |
Starch-binding domain |
Cyclodextrin glycosyltransferase, C-terminal domain |
Alkalophilic bacillus, sp. 1011 |
1pam |
a:583-686
|
0.785 |
R1D |
 |
d1erv__ 20%i.d. |
105 |
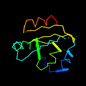
|
6.05 |
1 | 0.85

|
Alpha and beta proteins (a/b) |
Thioredoxin fold |
Thioredoxin-like |
Thioltransferase |
Thioredoxin |
Human (Homo sapiens) |
1erv |
-
|
0.500 |
R1D |
 |
d1a3wa1 20%i.d. |
99 |
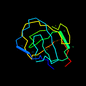
|
6.33 |
1 | 1.00
|
All beta proteins |
Pyruvate kinase beta-barrel domain |
Pyruvate kinase beta-barrel domain |
Pyruvate kinase beta-barrel domain |
Pyruvate kinase |
Baker's yeast (Saccharomyces cerevisiae) |
1a3w |
a:88-188
|
0.968 |
R1D |
 |
d1ebma2 19%i.d. |
124 |
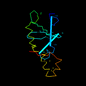
|
6.38 |
1 | n/a |
Alpha and beta proteins (a+b) |
TBP-like |
TATA-box binding protein-like |
DNA repair glycosylase, N-terminal domain |
8-oxoguanine glycosylase |
Human (Homo sapiens) |
1ebm |
a:9-135
|
0.853 |
3D |
 |
d1kb5b_ 24%i.d. |
117 |

|
7 |
0.562

| 0.42

|
All beta proteins |
Immunoglobulin-like beta-sandwich |
Immunoglobulin |
V set domains (antibody variable domain-like) |
T-cell antigen receptor |
Mouse (Mus musculus), beta-chain |
1kb5 |
b:
|
0.879 |
R1D |
 |
d1f13a3 19%i.d. |
101 |
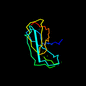
|
7.45 |
1 | 1.00
|
All beta proteins |
Immunoglobulin-like beta-sandwich |
Transglutaminase |
Transglutaminase |
Coagulation factor XIII, two C-terminal domains |
Human (Homo sapiens), blood |
1f13 |
a:628-728
|
0.000 |
3D |
 |
c1lvxa_ 23%i.d. |
74 |

|
7.77 |
1 | 1.00
|
All alpha proteins |
Binding domain of Skn-1 |
Binding domain of Skn-1 |
homol. to d1sknp_ |
- |
- |
1lvx |
a
|
0.000 |
R1D |
 |
c2fcba_ 10%i.d. |
173 |
|
7.87 |
0.85

| 0.57

|
not in SCOP 1.53 |
PDB header: immune system. |
Chain: A: PDB Molecule:fc gamma riib;. |
. |
PDBTitle: human fc gamma receptor iib ectodomain (cd32)
.
| - |
2fcb |
a
|
0.000 |
1D |