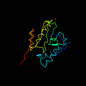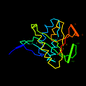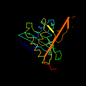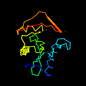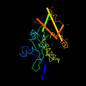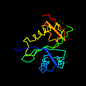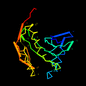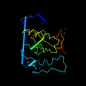| View Alignment |
Fold Library |
Template Length |
Model
| PSSM E_value |
SAWTED E-value |
Biotext |
Class |
Fold |
SuperFamily |
Family |
Protein |
Species |
ID |
Region |
CS |
PSSM Type
|
 |
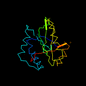
d1qsma_ 18%i.d. |
150 |
|
0.0998 |
0.376

| 0.16

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Histone acetyltransferase HPA2 |
Baker's yeast (Saccharomyces cerevisiae) |
1qsm |
a:
|
0.972 |
R1D |
 |
c1i1da_ 17%i.d. |
156 |

|
0.234 |
0.439

| 0.19

|
not in SCOP 1.53 |
PDB header: transferase. |
Chain: A: PDB Molecule:glucosamine-phosphate n-acetyltransferase;. |
. |
PDBTitle: crystal structure of yeast gna1 bound to coa and glnac-6p
.
| - |
1i1d |
a
|
0.000 |
R1D |
 |
c1ghea_ 14%i.d. |
166 |

|
0.47 |
1 | n/a |
not in SCOP 1.53 |
PDB header: transferase. |
Chain: A: PDB Molecule:acetyltransferase;. |
. |
PDBTitle: crystal structure of tabtoxin resistance protein complexed2 with an acyl coenzyme a
.
| - |
1ghe |
a
|
0.000 |
R1D |
 |
c1on0a_ 18%i.d. |
152 |
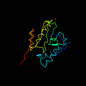
|
0.481 |
0.133

| n/a |
not in SCOP 1.53 |
PDB header: structural genomics, unknown function. |
Chain: A: PDB Molecule:yycn protein;. |
. |
PDBTitle: crystal structure of putative
acetyltransferase (yycn) from2 bacillus subtilis, northeast structural
genomics3 consortium target sr144 . | - |
1on0 |
a
|
0.000 |
R1D |
 |
d1cjwa_ 13%i.d. |
166 |
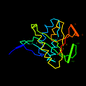
|
0.593 |
1 | 0.22

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Serotonin N-acetyltranferase |
Sheep (Ovis aries) |
1cjw |
a:
|
1.000 |
1D |
 |
c1mk4a_ 15%i.d. |
157 |

|
0.916 |
1 | n/a |
not in SCOP 1.53 |
PDB header: structural genomics, unknown function. |
Chain: A: PDB Molecule:hypothetical protein yqjy;. |
. |
PDBTitle: structural genomics, hypothetical protein yqjy from2 bacillus subtilis
.
| - |
1mk4 |
a
|
0.000 |
R1D |
 |
c1nsla_ 13%i.d. |
173 |

|
0.935 |
0.395

| n/a |
not in SCOP 1.53 |
PDB header: transferase. |
Chain: A: PDB Molecule:probable acetyltransferase;. |
. |
PDBTitle: crystal structure of probable acetyltransferase
.
| - |
1nsl |
a
|
0.000 |
R1D |
 |
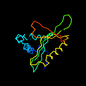
d1bo4a_ 15%i.d. |
137 |
|
1 |
0.461

| 0.26

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Aminoglycoside 3-N-acetyltransferase |
Serratia marcescens |
1bo4 |
a:
|
0.936 |
1D |
 |
d1b87a_ 13%i.d. |
181 |
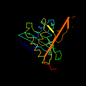
|
1.32 |
0.524

| 0.31

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Aminoglycoside 6'-N-acetyltransferase |
Enterococcus faecium |
1b87 |
a:
|
0.984 |
R1D |
 |
c1q2ya_ 23%i.d. |
140 |
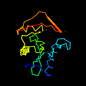
|
1.46 |
1 | n/a |
not in SCOP 1.53 |
PDB header: unknown function. |
Chain: A: PDB Molecule:similar to hypothetical proteins;. |
. |
PDBTitle: crystal structure of the
protein yjcf from bacillus2 subtilis: a member of the gcn5-related
n-acetyltransferase3 superfamily fold . | - |
1q2y |
a
|
0.000 |
R1D |
 |
d1qsta_ 11%i.d. |
160 |

|
4.02 |
1 | n/a |
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Catalytic domain of GCN5 histone acetyltransferase |
Tetrahymena thermophila |
1qst |
a:
|
0.984 |
R1D |
 |
c1m4da_ 16%i.d. |
181 |
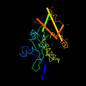
|
5.45 |
0.659

| n/a |
not in SCOP 1.53 |
PDB header: transferase. |
Chain: A: PDB Molecule:aminoglycoside 2'-n-acetyltransferase;. |
. |
PDBTitle: aminoglycoside 2'-n-acetyltransferase from mycobacterium2 tuberculosis-complex with coenzyme a and tobramycin
.
| - |
1m4d |
a
|
0.000 |
R1D |
 |
d1cm0a_ 10%i.d. |
162 |
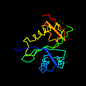
|
6.92 |
1 | n/a |
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Histone acetyltransferase domain of P300/CBP associating factor, PCAF |
Human (Homo sapiens) |
1cm0 |
a:
|
0.984 |
3D |
 |
c1p0ha_ 18%i.d. |
290 |

|
7.99 |
0.382

| n/a |
not in SCOP 1.53 |
PDB header: transferase. |
Chain: A: PDB Molecule:hypothetical protein rv0819;. |
. |
PDBTitle: crystal structure of rv0819 from mycobacterium tuberculosis2 mshd-mycothiol synthase coenzyme a complex
.
| - |
1p0h |
a
|
0.000 |
R1D |
 |
d1ygha_ 12%i.d. |
164 |
|
9.62 |
1 | n/a |
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Catalytic domain of GCN5 histone acetyltransferase |
Baker's yeast (Saccharomyces cerevisiae) |
1ygh |
a:
|
1.000 |
R1D |
 |
d1nmta1 9%i.d. |
165 |
|
27.5 |
0.57

| 0.02

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-myristoyl transferase, NMT |
N-myristoyl transferase, NMT |
Yeast (Candida albicans) |
1nmt |
a:60-224
|
0.900 |
3D |
 |
d2nmta2 12%i.d. |
237 |
|
27.9 |
0.692

| 0.02

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-myristoyl transferase, NMT |
N-myristoyl transferase, NMT |
Baker's yeast (Saccharomyces cerevisiae) |
2nmt |
a:219-455
|
0.971 |
R1D |
 |
d1nmta2 13%i.d. |
227 |
|
44.9 |
0.57

| 0.02

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-myristoyl transferase, NMT |
N-myristoyl transferase, NMT |
Yeast (Candida albicans) |
1nmt |
a:225-451
|
0.915 |
3D |
 |
c1k4ja_ 9%i.d. |
193 |
|
48 |
1 | 1.00
|
not in SCOP 1.53 |
PDB header: ligase. |
Chain: A: PDB Molecule:acyl-homoserinelactone synthase esai;. |
. |
PDBTitle: crystal structure of the acyl-homoserinelactone synthase2 esai complexed with rhenate
.
| - |
1k4j |
a
|
0.000 |
R1D |
 |
d1bob__ 16%i.d. |
306 |
|
52.5 |
0.591

| 0.12

|
Alpha and beta proteins (a+b) |
Acyl-CoA N-acyltransferases (Nat) |
Acyl-CoA N-acyltransferases (Nat) |
N-acetyl transferase, NAT |
Histone acetyltransferase HAT1 |
Baker's yeast (Saccharomyces cerevisiae) |
1bob |
-
|
0.966 |
3D |