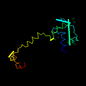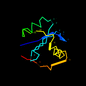| View Alignment |
Fold Library |
Template Length |
Model
| PSSM E_value |
SAWTED E-value |
Biotext |
Class |
Fold |
SuperFamily |
Family |
Protein |
Species |
ID |
Region |
CS |
PSSM Type
|
 |
c1iv0a_ 28%i.d. |
98 |
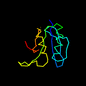
|
1.81e-06 |
1 | n/a |
not in SCOP 1.53 |
PDB header: structural genomics, unknown function. |
Chain: A: PDB Molecule:hypothetical protein;. |
. |
PDBTitle: solution structure of the yqgf-family protein (n-terminal2 fragment)
.
| - |
1iv0 |
a
|
0.000 |
R1D |
 |
d1hjra_ 17%i.d. |
158 |

|
0.163 |
1 | n/a |
Alpha and beta proteins (a/b) |
Ribonuclease H-like motif |
Ribonuclease H-like |
RuvC resolvase |
RuvC resolvase |
Escherichia coli |
1hjr |
a:
|
0.500 |
1D |
 |
c1m5ta_ 15%i.d. |
123 |

|
1.77 |
1 | n/a |
Alpha and beta proteins (a/b) |
Flavodoxin-like |
CheY-like |
homol. to d1ntr__ |
- |
- |
1m5t |
a
|
0.000 |
R1D |
 |
d1a2oa1 19%i.d. |
140 |

|
2.34 |
1 | n/a |
Alpha and beta proteins (a/b) |
Flavodoxin-like |
CheY-like |
CheY-related |
Methylesterase CheB, N-terminal domain |
Salmonella typhimurium |
1a2o |
a:1-140
|
0.596 |
R1D |
 |
d1b9da_ 17%i.d. |
143 |

|
2.84 |
1 | n/a |
Alpha and beta proteins (a/b) |
Ribonuclease H-like motif |
Ribonuclease H-like |
Retroviral integrase |
HIV-1 integrase, catalytic domain |
Human immunodeficiency virus, type 1 |
1b9d |
a:
|
0.500 |
R1D |
 |
c1ioia_ 22%i.d. |
208 |

|
3.13 |
1 | n/a |
Alpha and beta proteins (a/b) |
Phosphorylase/hydrolase-like |
Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
homol. to d1a2za_ |
- |
- |
1ioi |
a
|
0.000 |
R1D |
 |
c1g17a_ 15%i.d. |
168 |
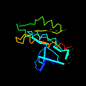
|
3.2 |
1 | n/a |
Alpha and beta proteins (a/b) |
P-loop containing nucleotide triphosphate hydrolases |
P-loop containing nucleotide triphosphate hydrolases |
homol. to d3raba_ |
- |
- |
1g17 |
a
|
0.000 |
R1D |
 |
d1b74a1 22%i.d. |
105 |
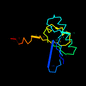
|
3.41 |
1 | n/a |
Alpha and beta proteins (a/b) |
ATC-like |
Glutamate racemase |
Glutamate racemase |
Glutamate racemase |
Bacteria (Aquifex pyrophilus) |
1b74 |
a:1-105
|
0.000 |
R1D |
 |
d1rvv1_ 23%i.d. |
154 |
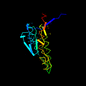
|
3.41 |
1 | n/a |
Alpha and beta proteins (a/b) |
beta-subunit of the lumazine synthase/riboflavin synthase complex |
beta-subunit of the lumazine synthase/riboflavin synthase complex |
beta-subunit of the lumazine synthase/riboflavin synthase complex |
beta-subunit of the lumazine synthase/riboflavin synthase complex |
Bacillus subtilis |
1rvv |
1:
|
0.000 |
R1D |
 |
d1fgs_1 16%i.d. |
111 |

|
3.44 |
1 | n/a |
Alpha and beta proteins (a/b) |
Glutamate ligase domain |
Glutamate ligase domain |
Folylpolyglutamate synthetase, C-terminal domain |
Folylpolyglutamate synthetase, C-terminal domain |
Lactobacillus casei |
1fgs |
297-425
|
0.000 |
R1D |
 |
c1mvoa_ 21%i.d. |
121 |

|
3.87 |
1 | n/a |
Alpha and beta proteins (a/b) |
Flavodoxin-like |
CheY-like |
homol. to d1ntr__ |
- |
- |
1mvo |
a
|
0.000 |
R1D |
 |
d1cxqa_ 10%i.d. |
143 |

|
5.19 |
1 | n/a |
Alpha and beta proteins (a/b) |
Ribonuclease H-like motif |
Ribonuclease H-like |
Retroviral integrase |
ASV integrase, catalytic domain |
Avian sarcoma virus, Rous sarcoma virus, Schmidt-Ruppin strain B |
1cxq |
a:
|
0.500 |
3D |
 |
c1gdta_ 16%i.d. |
183 |

|
5.54 |
1 | n/a |
not in SCOP 1.53 |
PDB header: complex (site-specific recombinase/dna). |
Chain: A: PDB Molecule:gamma-delta resolvase; 1gdt 6. |
. |
PDBTitle: crystal structure of a
site-specific recombinase, 1gdt 32 gamma-delta resolvase
complexed with a 34 bp cleavage site 1gdt 4 . | - |
1gdt |
a
|
0.000 |
R1D |
 |
c1o4va_ 18%i.d. |
169 |
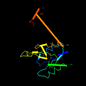
|
6.01 |
1 | n/a |
not in SCOP 1.53 |
PDB header: lyase. |
Chain: A: PDB Molecule:phosphoribosylaminoimidazole carboxylase. |
. |
PDBTitle: crystal structure of phosphoribosylaminoimidazole2 carboxylase (pure) (tm0446) from thermotoga maritima at3 1.77 a resolution
.
| - |
1o4v |
a
|
0.000 |
R1D |
 |
d1a5z_1 13%i.d. |
140 |
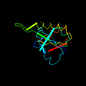
|
6.09 |
1 | n/a |
Alpha and beta proteins (a/b) |
NAD(P)-binding Rossmann-fold domains |
NAD(P)-binding Rossmann-fold domains |
Lactate & malate dehydrogenases, N-terminal domain |
Lactate dehydrogenase |
Thermotoga maritima |
1a5z |
22-163
|
0.500 |
R1D |
 |
c1j24a_ 14%i.d. |
133 |

|
6.18 |
1 | n/a |
not in SCOP 1.53 |
PDB header: hydrolase. |
Chain: A: PDB Molecule:atp-dependent rna helicase, putative;. |
. |
PDBTitle: crystal structure of archaeal xpf/mus81 homolog, hef from2 pyrococcus furiosus, nuclease domain, ca cocrystal
.
| - |
1j24 |
a
|
0.000 |
R1D |
 |
d1lam_1 15%i.d. |
159 |
|
6.56 |
1 | n/a |
Alpha and beta proteins (a/b) |
Leucine aminopeptidase, N-terminal domain |
Leucine aminopeptidase, N-terminal domain |
Leucine aminopeptidase, N-terminal domain |
Leucine aminopeptidase, N-terminal domain |
Bovine (Bos taurus) |
1lam |
1-159
|
0.500 |
R1D |
 |
c1sfe__ 17%i.d. |
165 |
|
6.73 |
1 | n/a |
not in SCOP 1.53 |
PDB header: dna-binding protein. |
Chain:NULL;: PDB Molecule:ada o6-methylguanine-dna methyltransferase;
. |
PDBTitle: ada o6-methylguanine-dna methyltransferase from escherichia2 coli
. |
- |
- |
1sfe |
-
|
0.000 |
R1D |
 |
d1tfr_1 16%i.d. |
123 |
|
6.85 |
1 | n/a |
All alpha proteins |
SAM domain-like |
5' to 3' exonuclease, C-terminal subdomain |
5' to 3' exonuclease, C-terminal subdomain |
T4 RNase H |
Bacteriophage T4 |
1tfr |
183-305
|
0.378 |
1D |
 |
d1sfe_2 21%i.d. |
81 |
|
7.46 |
1 | n/a |
Alpha and beta proteins (a/b) |
Ribonuclease H-like motif |
Methylated DNA-protein cysteine methyltransferase domain |
Methylated DNA-protein cysteine methyltransferase domain |
Ada DNA repair protein |
Escherichia coli |
1sfe |
12-92
|
0.500 |
R1D |