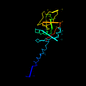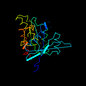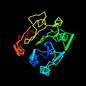| View Alignment |
Fold Library |
Template Length |
Model
| PSSM E_value |
SAWTED E-value |
Biotext |
Class |
Fold |
SuperFamily |
Family |
Protein |
Species |
ID |
Region |
CS |
PSSM Type
|
 |
c1i9ya_ 25%i.d. |
336 |

|
6.46e-16 |
0.312

| 0.33

|
not in SCOP 1.53 |
PDB header: hydrolase. |
Chain: A: PDB Molecule:phosphatidylinositol phosphate phosphatase;. |
. |
PDBTitle: crystal structure of inositol polyphosphate 5-phosphatase2 domain (ipp5c) of spsynaptojanin
.
| - |
1i9y |
a
|
0.000 |
R1D |
 |
d1ako__ 14%i.d. |
268 |

|
0.0183 |
0.943

| 0.67

|
Alpha and beta proteins (a+b) |
DNase I-like |
DNase I-like |
DNase I-like |
DNA-repair enzyme exonuclease III |
Escherichia coli |
1ako |
-
|
0.656 |
1D |
 |
d2dnja_ 13%i.d. |
253 |
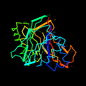
|
1.82 |
0.969

| 0.78

|
Alpha and beta proteins (a+b) |
DNase I-like |
DNase I-like |
DNase I-like |
Deoxyribonuclease I |
Bovine (Bos taurus) |
2dnj |
a:
|
0.804 |
R1D |
 |
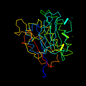
d1bix__ 13%i.d. |
275 |
|
1.97 |
0.923

| 0.62

|
Alpha and beta proteins (a+b) |
DNase I-like |
DNase I-like |
DNase I-like |
DNA repair endonuclease Hap1 |
Human (Homo sapiens) |
1bix |
-
|
0.758 |
3D |
 |
c1j6wa_ 14%i.d. |
155 |
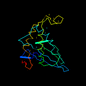
|
18.3 |
0.921

| 1.00
|
not in SCOP 1.53 |
PDB header: signaling protein. |
Chain: A: PDB Molecule:autoinducer-2 production protein luxs;. |
. |
PDBTitle: crystal structure of haemophilus influenzae luxs
.
| - |
1j6w |
a
|
0.000 |
1D |
 |
d1ei1a1 13%i.d. |
172 |
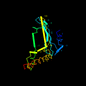
|
22.5 |
0.92

| 1.00
|
Alpha and beta proteins (a+b) |
Ribosomal protein S5 domain 2-like |
Ribosomal protein S5 domain 2-like |
DNA gyrase/MutL, second domain |
DNA gyrase B |
Escherichia coli |
1ei1 |
a:221-392
|
0.500 |
3D |
 |
c1ekea_ 14%i.d. |
219 |

|
23.6 |
0.973

| 0.81

|
not in SCOP 1.53 |
PDB header: hydrolase. |
Chain: A: PDB Molecule:ribonuclease hii;. |
. |
PDBTitle: crystal structure of class ii ribonuclease h (rnase hii)2 with mes ligand
.
| - |
1eke |
a
|
0.000 |
R1D |
 |
d1ceqa2 10%i.d. |
166 |
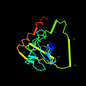
|
25.5 |
1 | 1.00
|
Alpha and beta proteins (a+b) |
Lactate & malate dehydrogenases, C-terminal domain |
Lactate & malate dehydrogenases, C-terminal domain |
Lactate & malate dehydrogenases, C-terminal domain |
Lactate dehydrogenase |
Malarial parasite (Plasmodium falciparum) |
1ceq |
a:164-329
|
0.573 |
R1D |
 |
d3nul__ 16%i.d. |
127 |
|
26.3 |
0.945

| 1.00
|
Alpha and beta proteins (a+b) |
Profilin-like |
Profilin (actin-binding protein) |
Profilin (actin-binding protein) |
Profilin (actin-binding protein) |
Mouse-ear cress (Arabidopsis thaliana) |
3nul |
-
|
0.754 |
1D |
 |
d1aa6_1 10%i.d. |
133 |
|
26.5 |
0.866

| 0.93

|
All beta proteins |
Double psi beta-barrel |
ADC-like |
Formate dehydrogenase/DMSO reductase, C-terminal domain |
Formate dehydrogenase H |
Escherichia coli |
1aa6 |
565-715
|
0.709 |
R1D |
 |
d1dyr__ 16%i.d. |
205 |

|
27 |
1 | 0.99

|
Alpha and beta proteins (a/b) |
Dihydrofolate reductases |
Dihydrofolate reductases |
Dihydrofolate reductases |
Dihydrofolate reductases, eukaryotic type |
Fungus (Pneumocystis carinii) |
1dyr |
-
|
0.800 |
3D |
 |
d1ldm_2 17%i.d. |
169 |

|
28 |
0.676

| 1.00
|
Alpha and beta proteins (a+b) |
Lactate & malate dehydrogenases, C-terminal domain |
Lactate & malate dehydrogenases, C-terminal domain |
Lactate & malate dehydrogenases, C-terminal domain |
Lactate dehydrogenase |
Dogfish (Squalus acanthias) |
1ldm |
161-329
|
0.683 |
1D |
 |
d1cipa2 13%i.d. |
195 |
|
29.2 |
0.881

| 0.93

|
Alpha and beta proteins (a/b) |
P-loop containing nucleotide triphosphate hydrolases |
P-loop containing nucleotide triphosphate hydrolases |
G proteins |
Transducin (alpha subunit) |
Rat (Rattus rattus) |
1cip |
a:32-60,a:182-347
|
0.500 |
3D |
 |
d1amoa3 12%i.d. |
160 |
|
29.4 |
0.949

| 0.94

|
Alpha and beta proteins (a/b) |
Ferredoxin reductase-like, C-terminal NADP-linked domain |
Ferredoxin reductase-like, C-terminal NADP-linked domain |
NADPH-cytochrome p450 reductase |
NADPH-cytochrome p450 reductase |
Rat (Rattus norvegicus) |
1amo |
a:519-678
|
1.000 |
3D |
 |
d8dfr__ 16%i.d. |
186 |

|
31.1 |
1 | 1.00
|
Alpha and beta proteins (a/b) |
Dihydrofolate reductases |
Dihydrofolate reductases |
Dihydrofolate reductases |
Dihydrofolate reductases, eukaryotic type |
Chicken (Gallus gallus) |
8dfr |
-
|
0.715 |
3D |
 |
d1d5ya3 14%i.d. |
169 |

|
31.2 |
0.94

| 1.00
|
Alpha and beta proteins (a+b) |
Probable bacterial effector-binding domain |
Probable bacterial effector-binding domain |
Rob transcription factor, C-terminal domain |
Rob transcription factor, C-terminal domain |
Escherichia coli |
1d5y |
a:122-294
|
0.000 |
1D |
 |
c1b08a_ 14%i.d. |
152 |
|
32.4 |
0.918

| 0.83

|
not in SCOP 1.53 |
PDB header: sugar binding protein. |
Chain: A: PDB Molecule:lung surfactant protein d;. |
. |
PDBTitle: lung surfactant protein d (sp-d) (fragment)
.
| - |
1b08 |
a
|
0.000 |
R1D |
 |
d1etu__ 14%i.d. |
177 |
|
32.6 |
0.857

| 1.00
|
Alpha and beta proteins (a/b) |
P-loop containing nucleotide triphosphate hydrolases |
P-loop containing nucleotide triphosphate hydrolases |
G proteins |
Elongation factor Tu (EF-Tu), N-terminal (G) domain |
Escherichia coli |
1etu |
-
|
0.500 |
3D |
 |
d1atza_ 8%i.d. |
184 |
|
33.2 |
0.933

| 0.84

|
Alpha and beta proteins (a/b) |
Integrin A (or I) domain |
Integrin A (or I) domain |
Integrin A (or I) domain |
von Willebrand factor A3 domain |
Human (Homo sapiens) |
1atz |
a:
|
0.785 |
3D |
 |
d1dfma_ 16%i.d. |
218 |
|
33.3 |
1 | n/a |
Alpha and beta proteins (a/b) |
Restriction endonuclease-like |
Restriction endonuclease-like |
Restriction endonuclease BglII |
Restriction endonuclease BglII |
Bacillus subtilis (synonym: Bacillus globigii) |
1dfm |
a:
|
0.500 |
3D |